INTRODUCTION
Transforming growth factor beta- 1 (TGF╬▓ 1) is a cytokine derived from various cells, including leukocytes, Kupffer cells and hepatic lipocytes1,2). It has regulatory roles in cell growth and differentiation1,2) and it has been reported to play a key regulatory role in the process of hepatic fibrogenesis by promoting the synthesis of extracellular matrix proteins, collagen and fibronectin4,5). It has also been suggested that TGF╬▓ 1 may have an important role in the development, growth or progression of tumors by the fact that it is upregulated in the cultured cell lines or tissues of several tumors6,7).
Genotype and serum titer of hepatitis C rirus(HCV) have been reported to affect the clinical presentations of patients with chronic hepatitis C in terms of the responsiveness to interferon therapy8ŌĆō10), progression of chronic hepatitis and occurrence of hepatocellular carcinoma (HCC)11ŌĆō14).
In this study, we were to evaluate whether different genotype or serum titer of causes any difference in the expression of serum TGF╬▓ 1. We also intended to examine the correlation between the up-regulation of TGF╬▓ 1 and the development or progression of HCC in patients with chronic hepatitis C.
PATIENTS AND METHODS
1. Patients
We studied 19 patients with chronic hepatitis and 18 with HCC associated with chronic HCV infection. All patients with chronic hepatitis had elevated serum aminotransferase values for more than 6 months, and also had HCV-RNA in their sera. Patients with serum HBsAg and those with a history of alcohol (over 40 g/day in males, 20 g/day in females over 10 years) or drug abuse were excluded. HCC was diagnosed histologically or based on the findings of hypervascular liver masses on CT scan with serum alpha-fetoprotein levels exceeding 400 ng/mL.
Their clinical records were reviewed for demographic and serum biochemical data (Table 1). The mean age of patients with HCC appeared to be 10 years older than those with chronic hepatitis. Also, serum albumin levels were lower and prothrombin time tended to be more prolonged in HCC than in chronic hepatitis, suggesting that the hepatocellular function of HCC patients may be more deteriorated compared to those with chronic hepatitis.
2. Genotyping and Quantitation of HCV-RNA
Total cellular RNA was extracted from serum by single-step acid guanidinium thiocyanate method with modifications15,16) using RNA STAT-60 (TEL-TEST ŌĆ£BŌĆØ, Friendswood, TX). Using products of reverse transcription polymerase chain reaction of 5ŌĆ▓ untranslated region (5ŌĆ▓UTR), HCV genotypes were determined by line probe reverse hybridization assay17,18) (INNO-LiPA HCV II, Innogenetics, Ghent, Belgium).
The amounts of HCV RNA in the sera were quantitated by branched DNA (bDNA) signal amplification assay19) (Quantiplex, Chiron Diagnostics, Emeryville, CA).
3. Measurement of Serum TGF╬▓ 1
Serum TGF╬▓ 1 levels were measured by enzyme linked immunosorbent assay (ELISA) using a commercial kit (TGF╬▓ 1 ELISA system, Promega, Madison, WI, USA).
4. Statistics
The values of serum HCV-RNA and TGF╬▓ 1 were transformed to logarithmic scale before analysis. The results represented as mean┬▒SD were compared by StudentŌĆÖs t test. Chi square test or FisherŌĆÖs exact test was used to evaluate the differences in proportion. SpearmanŌĆÖs rank correlation coefficient tested the correlation between the variables. p value < .05 was considered as significant.
RESULTS
1. Genotypes of HCV
Among 37 patients with chronic hepatitis or HCC associated with chronic HCV infection, 26 (70%) were infected with HCV of genotype 1b, 8 (22%) with non- 1b and 3 (8%) with both. HCV genotypes of patients with HCV-related HCC were quite similar to those with chronic hepatitis (genotype 1b: 67% vs. 74%; p=NS).
2. Serum Titers of HCV-RNA
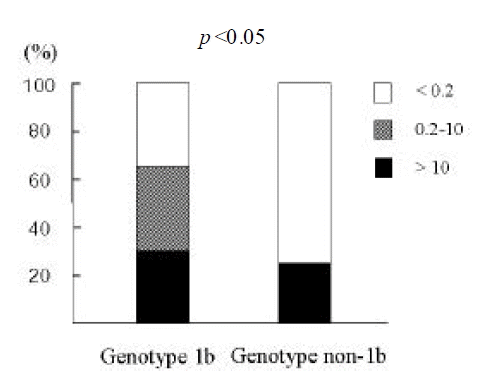
In 15 out of 37, the serum titers of HCV RNA were below 0.2 MEq/mL, which is the cut-off value of bDNA assay. The serum titers of HCV RNA tended to be higher in patients with HCC than those with chronic hepatitis (log value, MEq/mL; 1.05┬▒0.98 vs. 1.61┬▒0.92), although it did not reach the statistical significance (p=0.06). Moreover, the proportion of patients with HCV RNA levels > 0.2 MEq/mL was higher in patients with HCC than in those with chronic hepatitis (78% vs. 42%; p < 0.05) (Figure 1). This trend was observed regardless of the genotype of HCV. The amount of serum HCV RNA tended to be higher in genotype 1b compared to genotype non- 1b (> 0.2 MEq/mL; 65% vs. 25%; p < 0.05) (Figure 2).
3. Expression of Serum TGF╬▓1
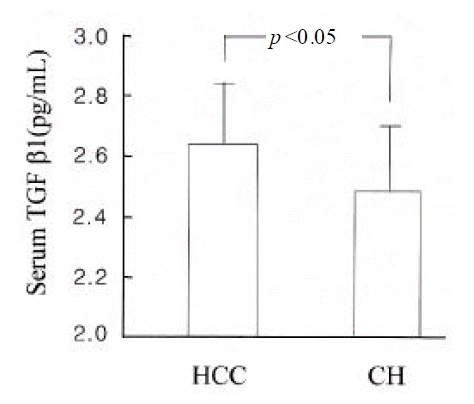
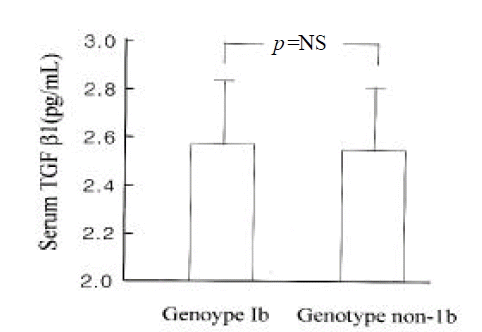
Serum TGF╬▓ 1 levels were higher in patients with HCC than in those with chronic hepatitis (log value, pg/mL; 2.48┬▒0.25 vs. 2.64┬▒0.22; p<0.05) (Figure 3). However, they were not different between genotype 1b and non- 1b (log value pg/mL; 2.56┬▒0.26 vs. 2.56 (0.23; p=NS) (Figure 4). Serum TGF?1 levels were not correlated with serum HCV-RNA titers or serum ALT levels (R=0.00 and R=0.01, respectively) (Figure 5).
DISCUSSION
TGF╬▓1 appeared to be overexpressed in patients with HCC compared to those with chronic hepatitis in this study. It suggests that TGF╬▓ 1 may have a role in the development or progression of HCC in patients with chronic hepatitis C. However, serum TGF╬▓1 level was not different among patients infected with various genotypes of HCV. Also, it was not correlated with serum HCV-RNA titers or ALT levels in patients with chronic HCV infection.
Our data indicated that higher serum titer of HCV-RNA may be more intimately related to genotype 1b as well as the development of HCV-associated HCC. This finding correlates with the previous reports111ŌĆō14) that high titer of serum HCV-RNA associated frequently with HCC in patients with chronic hepatitis C, and that, especially, genotype 1b had higher serum HCV-RNA level and induced more rapid progression to cirrhosis and HCC. However, in our data, regardless of its genotype, serum HCV-RNA titer of patients with HCC was higher compared to chronic hepatitis, suggesting that the serum titer of HCV-RNA may be more important than the genotype of HCV in the development of HCC.
Interestingly, there was no correlation between the amounts of TGF╬▓ 1 and the levels of HCV-RNA in the sera of patients with HCC despite the fact thatboth of them were higher than in those of chronic hepatitis. This finding indicates that TGF╬▓ 1 may exert effects on the process of hepatocarcinogenesis different from those of active HCV replication in patients with chronic hepatitis C. A previous report20) suggested that TGF╬▓ 1 may have much more of a role in the early stage of hepatocarcinogenesis. On the other hand, mutation of p53 gene was more frequently found in the poorly differentiated, multinodular and large-sized HCCs21). Recently, there have been some reports22,23) that vascular endothelial growth factor (VEGF) is also associated with the poorly differentiated or advanced stage of HCC. Thus, various growth factors, including TGF╬▓ 1 and VEGF or genetic alterations such as p53 mutation may have a role in the development or progression of HCC at the different stages of carcinogenesis. Active replication of HCV has been reported to activate various immunocytes in liver tissue followed by the release of various growth factors, which are involved in the transformation of hepatocytes or the progression of tumor cells24ŌĆō27). Clinically, HCV-associated HCC has been known to be different from the HBV-associated tumor in terms of the mean age of patients, the type of tumors and the pathology of the underlying liver28). Thus, it is suggested that the effects of HCV on the alteration of tumor-related gene and the release of various growth factors may be quite different from HBV29ŌĆō31). However, the exact role of HCV in the process of development of HCC remains to be clarified.
Our data also showed that serum TGF╬▓ 1 level was variable even in cases with HCC but we could not evaluate whether it is overexpressed in a certain type of tumor or in a specific clinical presentation of HCC patients because of the small number of subjects. The difference of TGF╬▓ 1 expression according to the clinical and oncological characteristics needs to be evaluated in the future. Furthermore, it is also necessary to clarify the exact mechanisms of TGF╬▓ 1 in hepatocarcinogenesis, including the expression of TGF╬▓ 1 mRNA in HCC and surrounding liver tissues, the changes of TGF╬▓ 1 receptor in cell membrane and the response of hepatocytes and interstitial cells to overexpressed TGF╬▓ 1.
In conclusion, TGF╬▓ 1 was overexpressed in HCC compared with chronic hepatitis C. However, it was not affected by HCV genotype or serum HCV-RNA titer. It is suggested that TGF╬▓ 1 may be associated with the malignant transformation of hepatocyte or the progression of HCV-associated HCC.





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print