Predictive Factors for Beneficial Response to Interferon-alfa Therapy in Chronic Hepatitis C
Article information
Abstract
Objectives:
Interferon is the only established teatment for chronic hepatitis C but the host-dependent or virus-related factors affecting the response rate to interferon therapy are not yet dear. The purpose of this study was to investigate the factors predictive of response to interferon-alfa therapy in chronic hepatitis C.
Methods:
Twenty-five consecutive patients with chronic hepatitis C were randomized to three regimens of interferon-alfa: group A (n=7, 3MU every day for 3 months), group B (n=8, 3MU every other day for 3 months) and group C (n=10, 3MU every other day for 6 months), We quantified serum HC RNA levels by competitive reverse transcription-polymerase chain reaction (RT-PCR)and performed HCV genotyping using type-specific primers deduced from the NS5 region of the HCV genome. We also attempted to identify which demographic, biochemical and histologic factors in addition to virus-related factors would significantly predict beneficial response to interferon by multivariate analysis.
Results:
Sustained responders were 8 (36.4%), nonsustained responders were 2 (9.1%) and nonresponders were 12 (54.5%) of 22 patients who had received complete therapy. The initial HCV RNA level (logarithmic transformed copy numbers per ml of serum)in sustained responders (5.75±0.39) was significantly lower than that of nonsustained responders (6.80±0.71)and nonresponders (6.70±0.52) (p<0.05).
In multivariate multiple logistic regression analysis, the serum HCV RNA level before therapy was only the independent predictor of a sustained response to interferon-alfa therapy (p=0.001).
Conclusions:
Serum HCV RNA level before therapy was the most useful predictor of a sustained response to interferon-alfa therapy for chronic hepatitis C.
INTRODUCTION
The hepatitis C virus (HCV) has known to be the major causative agent of post-transfusion hepatitis, since the cloning of the HCV genome was performed1,2). It is reported that about 50% of patients with HCV infection develop chronic hepatitis, and 10–25% of these cases will eventually progress to liver cirrhosis of hepatocellular carcinoma3).
Interferon (IFN) is considered the only approved therapy for chronic hepatitis C to date4–6), but the large number of relapses after cessation of therapy remains a problem.
Recently, many factors influencing the response to IFN therapy have been reported7–12). Among these, the HCV genotype and serum HCV RNA level of viral chracteristics have been recognized more specific than any other predictive factor10–12).
The purpose of this study was to evaluate clinical, biochemical and virological factors that would predict response to IFN-alfa therapy by univariate or multivariate analysis.
MATERIALS AND METHODS
1. Patients
Twenty five consecutive patients (16 men and 9 women) with chronic hepatitis C, with a mean age of 47 years (range: 23–58 years), were included in the study. All had persistently elevated serum alanine aminotransferase (ALT) levels of at least twice the upper normal limit of the normal range for at least 6 months, histological evidence of chronic hepatitis (chronic active hepatitis in 23 patients, chronic persistent hepatitis in 2 patients) and seropositivity for both anti-HCV [second generation enzyme immuno assay (EIA), Abbott Laboratories, North Chicago, IL, USA] and HCV RNA [reverse transcription-polymerase chain reaction (RT-PCR)]. Criteria for exclusion included evidence of infection with hepatitis A or B virus, decompensated liver disease, pregnancy or lactation, the presence of any serious medical or psychiatric illness and evidence of other forms of liver disease. Informed consent for participation was obtained from all patients before treatment.
Clinical and laboratory evaluation were performed monthly before and during treatment and then monthly between 6 and 36 months after treatment was completed.
2. IFN-alfa Therapy
Patients were randomly assigned to receive recombinant IFN alfa-2a (Intermax alpha; Lucky Biotec. Taejeon, Korea)according to three different protocols: group A (n=7), 3 million units (MU), subcutaneously (s. c.), every day for 3 months; group B (n=8), 3MU, s. c., every other day for 3 months; group C (n=10), 3 MU, s. c., every other day for 6 months.
3. Serological Assays
Serum anti-HCV was tested by a second generation commercial EIA kit and assayed before and at the end of treatment.
Oligonucleotide primers for detection, genotyping and quantitation of HCV RNA were synthesized by a DNA synthesizer (Pharmacia, LKB, Uppsala, Sweden).
4. HCV RNA Detection
HCV RNA was extracted from 100 μl of serum, as described previously13), and extracted RNA was precipitated with isopropanol and washed with ethanol. The HCV RNA pellet was dissolved in diethylpyrocarbonate-treated distilled water (DEPC-H20) and stored until use. Synthesis of cDNA from HCV RNA samples was carried out at 37°C for 1hour with 20 units of Moloney Murine Leukemia Virus reverse transcriptase (MMLV-RT, GIBCO BRL, Gaithersburg, MD. USA). The cDNA product was amplified on two stage PCR. The two sets of primer used for RT-PCR were derived from 5′-untranslated region (5′-UTR) of HCV genome14). The sequences were 5′-CTG TGA GGA ACT ACT GTC TT-3′ (sense, nucleotides 28 to 47) and 5′-CTG TGA GGA ACT ACT GTC TT-3′ (antisense, nucleotides 229 to 248) as an outer primer set, and 5′-TTC ACG CAG AAA GCG TC TAG-3′ (sense, nucleotides 46 to 65) and 5′-GTT GAT CCA AGA AAG GAC CC-3′ (antisense, nucleotides 171 to 190) as an inner primer set.
5. Genotyping
After extraction of HCV RNA and synthesis of cDNA as described above, the double stage PCR were performed with the four genotype-specific oligonucleotide primers described by Enomoto et al15). Kato et al.16,17) Chayama et al18) that were deduced from the nucleotide sequences in the NS5 region. The sequences of the universal primers for four genotypes used in the first round PCR were 5′-TGG GGA TCC CGT ATG ATA CCC GCT GCT TTGA-3′ (sense, corresponding to nucleotides 8230 to 8260 of HCV-J 16)) and 5′-GGC GGA ATT CCT GGT CAT AGC CTC CGT GAA-3′ (antisense, corresponding to nucleotides 8601 to 8630 of HCV-J 16)). The sequences of the primers in the second round PCR for HCV-US, -J, -K2a and -K2b, which are HCV genotypes 1a, 1b, 2a and 2b, respectively, according to the nomenclature of a recently proposed system 19), were 5′-CGA CAT CCGT ACG GAG GAGG-3′ (sense) and 5′-CAG GCT GCC CGG GCC TTG AT-3′ (antisense) for HCV genotype 1a, 5′-TGA CAT CCG TGT TGA GGA GT-3′ (sense)and 5′-CGG GCC GCA CAG GCC TCC AA-3′ (antisense) for genotype 1b, 5′-TAT GTT CAA CAG CAA GGG CCA GA-3′ (sense) and 5′-CCT GGT CAT AGC CTC CGT GAA-3′ (antisense) for genotype 2a, and 5′-CCT GGT CAT AGC CTC CGT GAA-3′ (sense) and 5′-CCTGGTCATAGCCTCCGTGAA - 3′ (antisense) for genotype 2b.
10μl of synthesized cDNA was amplified on two stage PCR as described previously20). The first PCR was carried out for 35 cycles of three steps, 1 min at 94°C, 1 min at 40°C and 1min at 72°C, respectively, and then 1 μl of the first PCR products was put into the prepared 4 each genotype specific microtube. The second PCR was carried out for 30 cycles of three steps, 1 min at 94°C, 45 seconds at 55°C, and 1 min at 72°C. The amplified DNA fragments were electrophoresed in 2% agarose gels with ethidium bromide staining and were observed under UV light.
6. Quantitative and Competitive Nested PCR
HCV RNA was quantified by a competitive RT-PCR assay using synthetic mutant HCV RNA as an internal template21,22). A mutant RNA was synthesized by means of site-directed mutagenesis and in vitro transcription.
A cDNA included 5′-UTR of HCV genome from the serum of HCV carrier was cloned into an M13mp18 vector system. After the above the M13mp 18-5′UTR plasmid was digested with restriction endonucleases, BamHI and Hind III (NEB, USA), and cloned again into pGEM4-z vector (pGEM-5′UTR). A deletion mutant (pGEM-5′UTRdel) was made by PCR. A mutant containing 80 base pairs deletion (pGEM-5′UTRdel)was digested Hind III, which served as templates for T7 RNA polymerase, and then the DNA template was removed using DNase I (Promega, RQ1 RNase free DNase I). The above mutant RNA transcripts were purified by phenol-chloroform extraction and ethanol precipitation and used as internal templates in quantitative competitive PCR. HCV RNA extraction using 50μl of serum was performed as described above.
An equal amount of sample RNA was put into a set into a set of microtubes that already had serially 10-fold diluted mutant RNA.
The sequences of primer used in the first PCR were HCV QC1 5′-CCA CCA TAG ATC ACT CCC CTGT-3′ (corresponding position 28 to 48 of HCV-II) as a sense primer and HCVL70, 5′-TTG AGG TTT AGG ATT CGT GCT CAT-3′ (corresponding position 343 to 366 of HCV-II) as an antisense primer.
The first PCR was carried out for 40 cycles of steps for 1min at 95°C, 1min at 55°C and 2min at 72°C, and then the second PCR was carried out for 25 cycles of steps for 1min at 94°C, 45seconds at 55°C and 1min at 72°C.
7. Efficacy Assessment
Complete response (CR) was defined as a normalization of serum ALT levels at the end of therapy. A sustained response (SR) was defined as the presence of persistently normal serum ALT levels and negative seroconversion of HCV RNA during the entire follow up (for at least six months) after CR. Nonsustained response (NSR) was of serum ALT levels became elevated again during follow up after CR. Patients were defined as nonresponders (NR) if serum ALT levels were persistently abnormal during or after treatment.
8. Statistical Methods
The Chi-square test or Fisher’s exact test was used for comparison between group frequencies. Univariate and multivariate analysis were used to establish which factors contributed to a sustained response to IFN therapy. We used SPSS (version 6.0) program for statistical analysis of all variables.
RESULTS
The patients included in this study were 25 patients (16 men and 9 women, with a mean age of 47, and an age range of 23 to 58), with seropositive anti-HCV, who had elevated serum ALT levels for at least 6months prior to the study. Four patients had a prior history of blood transfusions and the remainder had HCV infection from unknown causes. Three of the 25 patients had side effects (thrombocytopenia, leukopenia, depression) that led to discontinuation of therapy.
Table 1 shows the demographic, laboratory and histological profiles of the 25 patients before treatment.
The 22 patients who completed IFN therapy were divided into three groups according to their response to administration of IFN: SR, 8 of 22 patients (36.4%); NSR, 2 of 22 patients (9.1%) and NR, 12 of 22 patients (54.5%). No significant differences were seen in age, sex, history of transfusion and mean ALT of the three groups. No demographic or laboratory finding significantly affected the sustained response to IFN as determined by univariate or multivariate analysis.
As regards the relationship between the histological findings and the response to IFN-alfa therapy, for the 21 patients with chronic acive hepatitis (CAH), SR totalled 7 patients (33.3%), NSR totaled (9.5%) and NR totalled 12 patients (57.1 %). The sole patient with chronic persistent hepatistis (CPH) was a SR (Table 1).
The responses according to different IFN-α regimens were not significantly different in each group (Table 2).
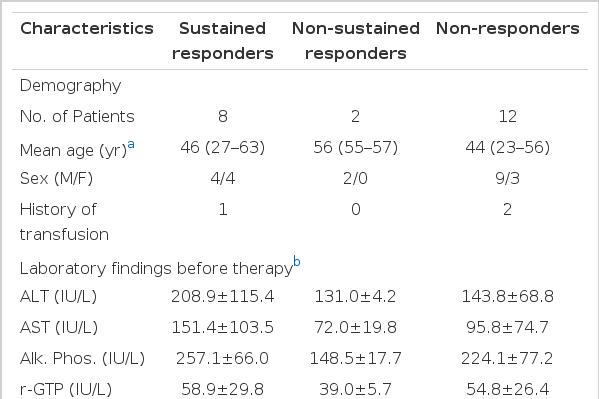
The results of HCV grnotyping shown in Table 3 and Fig. 1. 14 of 22 patients (63.6%) were genotype 1b, 7 of 22 patients (31.8%) were HCV 2a type and 1 patient had an undetermined type. In response to IFN-α according to HCV genotype, 5 (35.7%), 2 (14.3%) and 7 (50.0%) of 14 patients with HCV 1b type showed SR, NSR and NR, respectively. Of 7 patients with HCV 2a type, 3 (42.9%) and 4(57.1%) had SR and NR, respectively. No significant correlation between response to IFN-α and genotype was seen.
Detection of each HCV genotype by reverse transcription-nested polymerase chain reaction using the genotype specific primers deduced from NS5 region of HCV genome.
The upper portion [A] shows HCV-J (1b) type while the lower portion[B] shows HCV-K2a (2a) type. The letter “L” indicates molecular weight size marker (123 bp).
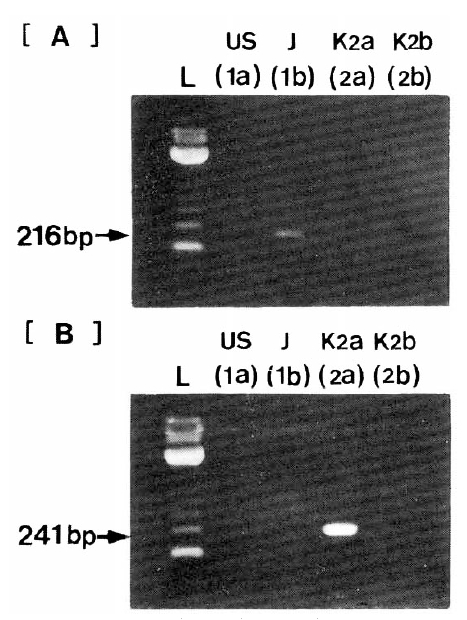
The quantitation by competitive RT-PCR was performed in all patients and the quantity of serum HCV RNA was determined in all patients, except one patient. Serum HCV RNA levels expressed as log10 (copy numbers of HCV RNA per ml serum) varied from 5.3 to 7.3. In the response to IFN-α according to serum HCV RNA levels before treatment, patients with SR had significantly lower serum HCV RNA levels (5.75±0.39) than those of patients with NSR or NR (6.80±0.71, 6.70±0.52, respectively) (p<0.05) (Fig. 3).
Comparisons of circulation HCV RNA quantity in each response group before IFN therapy. SR (•), stustained responder; NSR (▪), nonsustained responders; NR (▴), nonresponders.
Bars express mean±S.D. Logarithms of serum HCV RNA quantity (mean±S.D) were 5.75±0.39 in SR, 6.80±0.71 in NSR and 6.70±0.52 in NR. Significant differences in serum HCV RNA quantity were seen between SR, and NSR and NR (p<0.05).
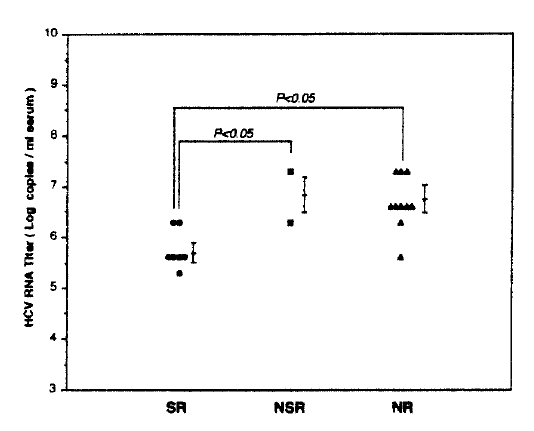
Patients were stratified to genotype 1b and -2a to see which is a more important predicor of response to IFN-α between serum HCV RNA levels before IFN therapy and genotype. In patients with genotype 1b, serum HCV RNA levels of SR, NSR and NR showed 5.95±0.40, 6.80±0.71 and 6.95±0.40 respectively. In two genotype groups serum, HCV RNA levels of SR were lower than those of patients with NSR or NR. However, there was no statistically significant difference between each group or between genotypes (Fig. 4).
Correlations between serum HCV RNA levels and HCV genotypes in three response groups before IFN therapy. SR (•), sustained responder; NSR (▪), nonsustained responders; NR (▴), nonresponders. No significant correlations in serum HCV RNA levels between each group or between genotypes were seen.
IFN was well tolerated in almost all patients and the side effects were generally not serious and the patients soon recovered. The common side effects were initially transient flu-like syndrome (100%), fatigue (48.0%), hair loss (44.0%), loss of appetite (40.0%), leukopeina (24.0%), thrombocytopenia (20.0%), emotional instability (12.0%) and thromboembolism (4.0%). Three of 25 patients who initially enrolled in the study stopped treatment due to serious side effects of marked thrombocytopenia and leukopenia, derpession and thromboembolism, respectively.
Multiple regression analysis in a stepwise method was performed to identify the variables (demographic, biochemical, histological, HCV genotype and HCV RNA levels) having significant correlation with sustained response to IFN therapy.
Serum HCV RNA level before therapy was only estimated to be independently predictive of a sustained response (p=0.001; 95% confidence intervals, 0.227–0.758).
DISCUSSION
IFN-α is known as the only established treatment for chronic hepatitis type C. The reason why not all patients respond to IFN therapy is not clear. However, patients characteristics, administration schedule of IFN therapy and molecular biological properties of HCV are thought to be some of the factors influencing the outcome of IFN therapy. Among them, shorter duration of disease, absence of cirrhosis, prolonged therapy with higher doses, a lower histologic activity index (HAI), HCV genotype 2 and lower level of serum HCV RNA have been associated with better response to IFN4,6,11–12,23–25).
In this study, demographic profiles, biochemical data and histological findings before IFN therapy had no important effect on the response rate to IFN, as reported previously26).
Although the shorter duration of disease is reported to predict a better response to IFN therapy12), we did not include it among possible variables that can influence the response to IFN therapy in this study, because the time of onset of HCV infection or hepatitis could not be fully evaluated by biochemical or virological studies in retrospective analysis.
Although higher initial doses and longer duration of IFN therapy showed higher long-term response rates4,6,12), compared to the standard 24 week, 3 MU TIW treatment schedule, the alternative treatment regimens for beneficial response to IFN have remained unclear. In our study, the sustained response rate according to different regimens of IFN-α was 42.9% of group A, 40.0% of group B and 30.0% of group C. However, no significant differences were seen in the response rate among these three groups. The response rate at treatment regimens with the same doses but with different durations between group A and C, was higher in group A. Moreover group B, with a regimen of shorter duration, had a better response rate than group C with regimen of longer duration. Thus, further extensive studies will be needed to clarify the relationship between the response rate to IFN and regimens of IFN including, doses and duration.
Sustained loss of serum HCV RNA was observed in the 8 SR but in none of the responders. This showed that HCV might be eradicated in 8 of the 22 patients (36.4%) treated with IFN therapy.
HCV genotype has been known to be associated with the response rate to IFN therapy and generally, genotype III (2a) has a better response to IFN than genotype II (1b)12,24). The reason why the HCV genotype influences the response rate to IFN is still unclear. Some researchers hypothesize it may be associated with the degree of heterogeneity of the hypervariable region (E2/NS1) within the HCV genome27). Our data showed that the sustained response rate was higher in patients with genotype 2a (42.9%) than in those with genotype 1b (35.7%), but this difference is not statistically significant.
We quantified HCV RNA in the sera from patients using a competitive RT-PCR assay with synthetic mutant HCV RNA as an internal template (competitor)21). The advantages of this method, compared to Kaneko’s method28), are that firstly, conserved 5′-UTR of HCV genome was used as a template and thus all genotypes of HCV could be detected; secondly, the efficiency of cDNA synthesis by reverse transcriptase could not influence the quantitiation of HCV RNA because synthetic mutant RNA was used as an internal template; and finally, any contamination could be easily found because synthetic mutant RNA was deleted in 80 base pairs of sequences. However, as competitive RT-PCR is a complicated and time consuming method, it is not available for mass clinical samples. Recently, an easier branched DNA signal amplification (bDNA) assay (Chiron, CA, USA) was developed for quantification of HCV RNA. This method will be commonly used in the future.
The initial titer of HCV RNA or HCV genotype by the previous reports25,26) was suggested as the most significant independent predictor of a sustained response to IFN therapy. In this study serum HCV RNA level before treatment was significantly lower in SR than in NSR and NR (P<0.05). These results suggested that interferon-α is effective for suppressing the replication of HCV. Also, multivariate multiple logistic regression showed that pretreatment HCV RNA level is a most significant independent predictor of a sustained response to IFN.
In the relationship between serum HCV RNA levels before therapy and HCV genotype in three groups, no significant correlation in levels of viremia between each group, or between genotypes, was seen. These results agree with Tsubota’s data26).
We conclude that serum HCV RNA level before therapy is the most important factor influencing the long-term response to IFN therapy.

Quantitation of HCV RNA by competitive polymerase chain reaction using the internal template, which was synthesized by site-directed mutagenesis and in vitro transcription, as control.
The upper bands (268 bp) indicate amplified DNA fragments derived from target HCV RNA, and the lower band (188 bp) indicate those from the internal template as competitor. At the equivalent point of signal intensities between the two bands, the amounts of target HCV RNA are the same as the copy numbers of the internal templates. The arrow on the lower portion indicates the point of titration of HCV RNA (4×104 copies/ml serum). The numbers above the oblique arrows on the upper portion are the copies of the internal templates. The letter “L” indicates molecular weight size marker (123bp).
CNC: negative control used in cDNA synthesis
PNC: negative control used in PCR process
Notes
This work was supported by grants from the Catholic Medical Center.