Cancer patients with neutropenic septic shock: etiology and antimicrobial resistance
Article information
Abstract
Background/Aims
Among patients with febrile neutropenia that developed after chemotherapy, high-risk patients, such as those having clinical instability or Multinational Association of Supportive Care in Cancer score of < 21, require hospitalization for intravenous empiric antibiotic therapy. Monotherapy with an anti-pseudomonal ß-lactam agent is recommended. Although many studies reported the microbial etiology of infections and resistant patterns of febrile neutropenia, the patients were not well characterized as having neutropenic septic shock. Therefore, this study aimed to determine the microbial spectrum of infections and resistance patterns of their isolates in patients with chemotherapy-induced neutropenic septic shock.
Methods
Data of adult patients diagnosed with neutropenic septic shock in the emergency department between June 2012 and December 2016 were extracted from a prospectively compiled septic shock registry at a single academic medical center. Thereafter, microbiological studies and antimicrobial susceptibility tests were conducted.
Results
In total, 109 bacteria were found in patients with neutropenic septic shock. Gram-negative bacteria were the predominant causative organisms (84, 77.1%). Moreover, 33 microorganisms (30.3%) were multidrug-resistant (MDR) bacteria with extended-spectrum ß-lactamase-producing Escherichia coli (17, 50%) being the commonest. The most commonly affected sites in patients with MDR bacterial infections were the gastrointestinal tract (45%) and unknown (43.5%). Approximately 48.5% of MDR bacteria were resistant to cefepime but not to piperacillin- tazobactam or carbapenem.
Conclusions
MDR bacteria were prevalent in patients with chemotherapy-induced neutropenic septic shock. Therefore, piperacillin-tazobactam or carbapenem may be considered as empiric antibiotics if MDR bacteria are suspected to be causative agents.
INTRODUCTION
Sepsis is a leading cause of mortality in patients with hematologic malignancies or solid tumors undergoing intensive cytotoxic chemotherapy [1]. Particularly, failure to initiate appropriate empiric therapy in patients with sepsis and septic shock is associated with a substantial increase in morbidity and mortality [2]. Neutropenia, defined as an absolute neutrophil count (ANC) of < 500/mm3, remains the most frequently occurring unfavorable consequence of chemotherapeutic agents. Administration of prompt, empiric, broad-spectrum, hospital-based, and parenteral antimicrobial therapy has become the standard of care for most patients with febrile neutropenia. Patients with febrile neutropenia that developed after chemotherapy with clinical instability or Multinational Association of Supportive Care in Cancer score of < 21, require hospitalization for intravenous (IV) empiric antibiotic therapy. Monotherapy with an anti-pseudomonal ß-lactam agent, i.e., cefepime, carbapenem, or piperacillin-tazobactam, is recommended. In particular, clinically unstable patients with febrile neutropenia may require a combination of broad-spectrum ß-lactam antibiotics and aminoglycoside to extend the antibacterial spectrum and to obtain a synergistic effect against some gram-negative bacteria [3,4]. However, the susceptibility and resistance of anticipated pathogens to antimicrobials varied over time globally, with a recent shift toward gram-negative bacteria in the bacterial epidemiology of hematological patients [5-7]. Moreover, the production of extended-spectrum ß-lactamases (ESBLs) and subsequent resistant gram-positive pathogens have drawn attention due to the spread of antimicrobial resistance.
Sepsis and septic shock are life-threatening conditions caused by a dysregulated immune response to infection and often lead to tissue and organ injures and death [8-10]. Although a few studies have evaluated the microbial etiology of patients with neutropenic sepsis, none of them have focused on neutropenic septic shock [1,11].
Therefore, a cohort study to determine the microbial spectrum of infections and resistance patterns of their isolates in patients with chemotherapy-induced neutropenic septic shock was conducted.
METHODS
Setting and study population
A single-center, retrospective, observational, registrybased study analyzing all patients diagnosed with neutropenic septic shock between June 1, 2012, and December 31, 2016, was conducted. The Asan Medical Center has an annual emergency department (ED) volume of approximately 110,000 patients and serves as a tertiary referral center in Seoul, Korea. All adult patients (aged ≥ 18 years) diagnosed with septic shock in the ED and treated with protocol-driven resuscitation bundle therapy since January 2010 were enrolled with their data prospectively collected in our septic shock registry.
All patients with septic shock were treated with protocol-driven resuscitation bundle therapy as indicated. In accordance with the surviving sepsis campaign, blood culture, empiric antibiotics, and initial lactate measurements were obtained within 3 hours of shock recognition. Blood cultures were performed at two or more different anatomical sites, in accordance with the standard practice. When an indwelling central venous catheter was present, one blood sample was collected from the catheter and the others were obtained from different peripheral veins. Additionally, specimens were obtained from the suspected lesions (e.g., sputum, urine, and pus), and further tests such as culture and polymerase chain reaction were performed. Patients with a “do not attempt resuscitation” status and those who were transferred from another hospital during the initial resuscitation were not included. In case of neutropenic septic shock, all patients were administered parenteral antib iotics such as extended-spectrum penicillin/β-lactamase inhibitors (piperacillin-tazobactam), fourth-generation cephalosporin (cefepime), cefazolin/ceftazidime combination, or meropenem.
All study participants provided informed consent, and the study design was approved by the appropriate Ethics Review Board of Asan Medical Center (2015-1253).
Data collection and definition
Demographic and clinical data including age, gender, previous medical history, symptoms, initial vital signs, site of infection, and laboratory values upon admission were retrieved from the septic shock registry. Septic shock was defined as refractory hypotension (mean arterial pressure of ≤ 70 mmHg) requiring vasopressors despite adequate fluid therapy or a blood lactate concentration of at least 4 mmol/L [12]. Neutropenia was defined as an ANC of < 0.5 × 109/L upon of ED admission.
The types of infections were classified into three groups: (1) unknown cause of infection, defined as the absence of any obvious infection upon clinical and radiological examination and a negative result upon microbiological examination; (2) clinically documented infection, defined as a either clinically or radiologically proven case of infection, but no evidence of the causative organism; and (3) microbiologically documented infection, defined as the presence of a bacterium in the infection site or a positive blood culture. Bacteremia caused by coagulase-negative Staphylococcus species (CoNS) was diagnosed after two positive results from two independent cultures, whereas bacteremia caused by other bacteria was diagnosed when they were positive in one culture test. Multidrug-resistance (MDR) was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories [13].
Statistical analysis
Continuous variables were expressed as mean ± standard deviation or medians with interquartile ranges if the assumption of a normal distribution was violated. Categorical variables were expressed as numbers and percentages. All tests in this study were two-sided, and a p value of < 0.01 was considered as statistically significant. All statistical analyses were performed using SPSS for Windows version 20.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
Patient characteristics
The study sample consisted of 133 patients with neutropenic septic shock: seven belonged to the “unknown cause” category, 33 exhibited “clinically documented infection,” and 93 exhibited “microbiologically documented infection.” The mean age of patients with microbiologically documented infection was 63.6 years, and 43.0% (n = 40) of them were women. The most common infection originated from the respiratory system (39.4%), followed by those affecting the gastrointestinal system (18.1%). Moreover, 17 patients (18.1%) in the “unknown origin” group were found to have no clinical or radiological cause of infection; however, the causative microorganisms were identified. The characteristics of each patient group are shown in Table 1.
Causative organisms in patients with neutropenic septic shock
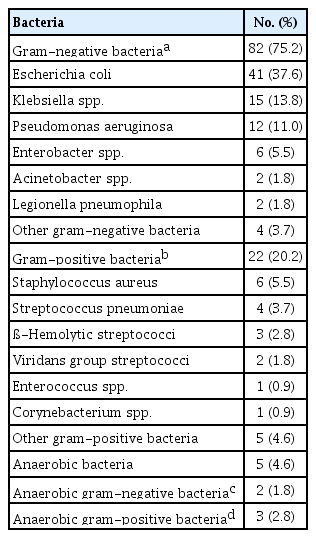
A total of 86 patients were diagnosed with bacterial infection, 15 with viral infection, and eight were coinfected with bacteria and viruses. Among the patients with bacterial infection, 69 were infected with one bacterium, and 17 patients were infected with two or more bacteria. On the test, a total of 109 bacteria were identified as causative organisms in patients with neutropenic septic shock. Among them, gram-negative bacteria, particularly Escherichia coli, were the most common causative organisms (77.1%). Gram-positive bacteria were also identified (22.9%), such as Staphylococcus aureus and S. pneumoniae (Table 2).
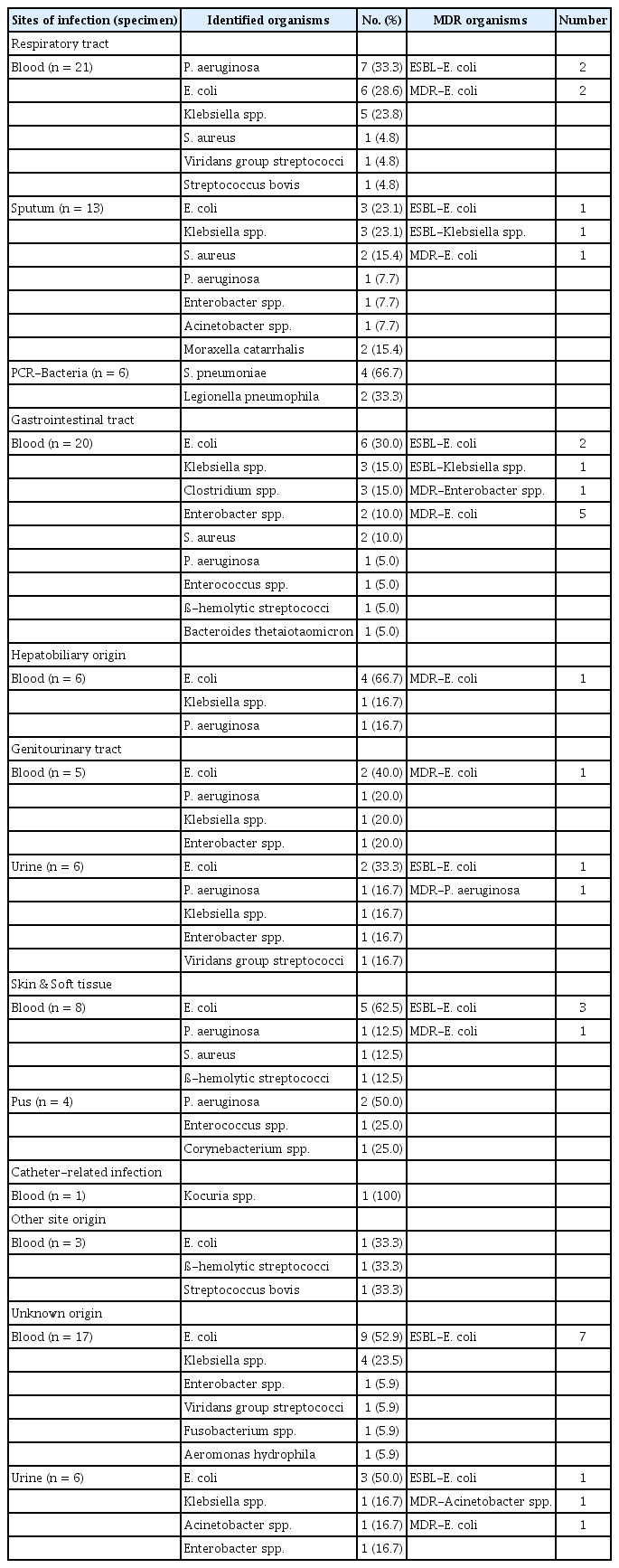
The infection sites were divided into eight groups. Table 3 summarizes the causative organisms identified according to infection site and specimen. In cases of respiratory tract infections, blood cultures showed that Pseudomonas aeruginosa (33.3%) were the most frequently identified causative agent. Blood cultures showed that E. coli were the most common causative organism in 30% of lesions affecting the gastrointestinal tract, 66.7% in the hepatobiliary, and 40% in the genitourinary tract. Gram-positive bacteria were most commonly found in patients with catheter-related infections. MDR organisms were observed in various sites and most frequently found in the gastrointestinal tract and unknown origin infections (Table 3).
MDR bacteria in patients with neutropenic septic shock
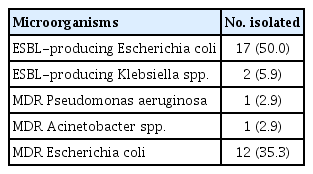
Antibiotic susceptibility tests were performed using blood or other specimens collected from the infected lesions, and the distribution of MDR bacteria were also examined. The results showed that 33 (30.3%) microorganisms satisfied the MDR definition used in this study. The most common MDR bacteria was ESBL-producing E. coli (50%), followed by MDR-E. coli (ESBL-negative, 35.3%), which was resistant to three or more antibiotics (Table 4).
MDR organisms were all susceptible to carbapenem and piperacillin-tazobactam. They were also susceptible to amikacin, an aminoglycoside recommended as a combination therapy for the management of complications. However, 48.5% and 72.7% of MDR organisms were resistant to cefepime and ceftazidime that were used as an anti-pseudomonal cephalosporin, respectively.
DISCUSSION
This study demonstrated that the proportion of MDR bacteria (30.3%) found in patients with chemotherapy-induced neutropenic septic shock is high. Furthermore, MDR bacteria were also found to be resistant to monotherapy with an anti-pseudomonal ß-lactam agent such as cefepime (48.5%).
The Infectious Diseases Society of America guidelines suggested that high-risk patients should be hospitalized for IV empiric antibiotic therapy, and monotherapy with anti-pseudomonal ß-lactam agents, i.e., cefepime, carbapenem (meropenem or imipenem-cilastatin), or piperacillin-tazobactam, is recommended. Although some observational data have suggested that highdose cefepime (e.g., 2 g every 8 hours) can be effective against ESBL-producing pathogens [14,15], most of the available evidence does not encourage its use. In a randomized trial of patients with urinary tract infection with ESBL-producing pathogens, treatment failure with cefepime was found to be higher than that with ertapenem or piperacillin-tazobactam [16]. Similarly, a retrospective study that used an MIC of ≤ 8 µg/mL as the cut-off point for cefepime susceptibility reported that cefepime was inferior to carbapenems against bacteremia caused by ESBL-producing Enterobacteriaceae [17]. Therefore, this evidence, along with the high prevalence of MDR pathogens resistant to cefepime observed in the current study, suggests that monotherapy with cefepime may not be suitable for the treatment of septic shock in patients with neutropenia.
According to the guidelines of administering empiric therapy for the treatment of febrile neutropenia in Korea, gram-negative bacteria were more frequently observed in studies conducted until the early 2000s. Among the gram-negative bacteria, E. coli is the most common, followed by P. aeruginosa and Klebsiella spp. Similarly, in our study, which was conducted after 2012, gram-negative organisms were also the most common causative organism, with E. coli as the most common pathogen. However, data regarding the antimicrobial susceptibility to pathogens in febrile neutropenia have been limited in Korea, and the reported resistance rates vary. The rate of third-generation cephalosporin-resistant E. coli was reported to be 0% to 7.0% [4].
In a study published in 2013, approximately 26% of bacteremia due to E. coli or Klebsiella pneumoniae were caused by ESBL-producing organisms [18]. In the current study, the proportion of MDR pathogens was high (approximately 35%), and all of them were gram-negative bacteria. In particular, approximately 34% of E. coli and Klebsiella spp., the most common causative agents of febrile neutropenia with septic shock, were ESBL-producing organisms. The high proportion of MDR pathogens in our study may be due to the regional and study population differences, and patients with neutropenic septic shock might have higher MDR pathogens than those with febrile neutropenia only.
Previous studies have demonstrated that primary bacteremia was the most common cause of infection in patients with neutropenia, followed by urinary tract infection and pneumonia [19]. The typical pathogen was slightly different for each type of infection, with P. aeruginosa and S. pneumoniae being the most common causative agents in infections originating from the lungs. Additional pathogens to be considered in cancer patients with neutropenia include gram-negative enteric organisms such as P. aeruginosa, Klebsiella spp., and E. coli [20]. The most common pathogens in infections originating in the abdomen and urogenital regions were E. coli, whereas soft tissue and central venous catheter-related infections were mainly caused by gram-positive bacteria. Among the patients belonging to the “unknown origin” group, the most common causative agents were noted to be CoNS and E. coli [1].
This study identified numerous causative organisms, which greatly varied according to the infection site. However, in contrast to the previous evidence, E. coli was most frequently identified in blood cultures from skin and soft tissue infections in this study. Skin and soft tissue infections caused by gram-negative bacteria, such as E. coli, are uncommon and present as primary skin infections with a cutaneous portal of entry [21]. Secondary bloodstream infections that originated from the digestive tract may also occur. However, the number of cases that have been previously reported was limited, and all of them were found to have occurred in immunocompromised patients [22].
The identification rate of MDR bacteria also differed according to the infection site. This variation in the proportion of MDR bacteria based on the infection site is crucial during the use of early empiric antibiotics. Therefore, antibiotics used for the treatment of septic shock in patients with neutropenia should be carefully selected, taking into consideration the increased rates of ESBL-producing organisms and the suspected infection site. Although gram-positive bacteria were also identified as causative organisms, MDR gram-positive bacteria such as methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus were not found in this study. Therefore, the use of vancomycin should be evaluated based on the patient’s condition and the risk factors present, such as previous infections, colonization with the organism, and treatment in a hospital with high endemic rates.
Our study had several limitations. First, this was an observational study using data from a prospective septic shock registry, and unmeasured confounding variables may exist. Second, although this study included a relatively large cohort of patients with neutropenic septic shock, the sample was too small to carry out specific analyses. Third, the data used in this study were collected from a single center; thus, the results should be cautiously interpreted in studies conducted in different populations and countries. Future studies should include controlled prospective trials that identify patients highly at risk of infection because of MDR pathogens.
In conclusion, MDR bacteria were prevalent (30.3%) in patients with chemotherapy-induced neutropenic septic shock, and 48.5% of them were resistant to cefepime. If MDR bacteria are suspected to be the causative agents, piperacillin-tazobactam or carbapenem may be considered as empiric antibiotics. Moreover, aminoglycoside may also be added to the initial regimen for the management of complications.
KEY MESSAGE
1. Failure to initiate appropriate empiric therapy in patients with sepsis and septic shock is associated with a substantial increase in morbidity and mortality.
2. Knowledge on the local epidemiology is essential in choosing the appropriate empiric antibiotic therapy.
3. In our study, multidrug-resistant bacteria were prevalent in patients with chemotherapy-induced neutropenic septic shock, and 48.5% of them were resistant to cefepime.
4. Piperacillin-tazobactam or carbapenem may be considered as empiric antibiotics in cancer patients with neutropenic septic shock.
Notes
No potential conflict of interest relevant to this article was reported.