HLA Antigens in Malay Patients with Systemic Lupus Erythematosus: Association with Clinical and Autoantibody Expression
Article information
Abstract
Background:
Studies have shown that certain genes within the major histocompatibility complex predispose to systemic lupus erythematosus (SLE) and may influence clinical and autoantibody expression. Thus, we studied the frequency of HLA-DR, -DQA, -DQB and -DPB alleles in ethnic Malays with SLE to determine the role of these genes in determining disease susceptibility and their association with clinical and immunological manifestations.
Methods:
Fifty-six Malay SLE patients were enrolled into the study. Demographic, clinical and immunological findings were obtained from medical records. HLA-DR, DQ and DP typing were done using modified PCR-RELP. Controls were from ethnically-matched healthy individuals.
Results:
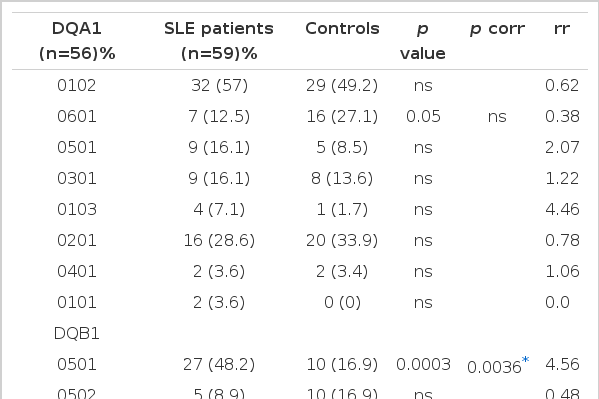
We found a strongly significant association of the DR2 and DQB1 *0501 and DQB1 *0601 (pcorr=0.03, rr=3.83, pcorr=0.0036, rr=4.56 and pcorr=0.0048 and rr=6.0, respectively). There was also a weak increase of DQB1 *0.201 and DPB1 *0.0901 with a weak decrease of DQA1 *0601 and DQB1 *0503 and *0301 which were not significant after corrections for multiple comparisons were made. There was a significant positive association of DR2 and DQB1 *0501 with renal involvement and DR8 with alopecia. A nonsignificant increase of DQB1 *0503 in patients with photosensitivity was noted. Significant autoantibody associations were also found: DQB1 *0601 with anti-Sm/RNP, DR2 with antiSSA (Ro)/SSB (La), and DR2, DQB1 *0501 and *0601 with antibodies to ds DNA. There was no specific DR, DQ or DP associations with age of disease onset (below 30 years or those at or above 30 years).
Conclusion:
Our data suggests the role of the HLA class II genes in conferring SLE susceptibility and in clinical and autoantibody expression
INTRODUCTION
Systemic lupus erythematosus is an unknown autoimmune disease which exhibits clinical and immunological heterogeneity. Genetic factors have been known to play some role in its pathogenesis. Though the genes involved in SLE have not yet been identified with certainty, most studies have focused on the genes within the major histocompatibility complex (mhc). It is also well known that the distribution of HLA antigen is different according to races. In Cavcasians, the most consistent associations that have been reported are those with HLA-B8, DR2, DR3, DQw1 and null alleles of the second and fourth component of the complement1–8). However, among American Blacks, association has been variously described with DR37), both DR2 and DR39) and DR7, though these too have not been consistent. The genes with in the mhc have also been found to influence clinical heterogeneity and immunological responses in these patients4,10–12). The purpose of this study was to determine the contribution of the HLA class II antigen in disease susceptibility and also to determine whether they play a role in expressing the clinical manifestations and immunological responses in SLE patients.
MATERIALS AND METHODS
The patient group consisted of 56 Malay patients with SLE, made up of 52 (93%) females and 4 (7%) males, giving a female: male ratio of 13:1. The patients are those attending the SLE Clinic of the National University Hospital of Malaysia and who met the American College of Rheumatology classification criteria for SLE (formerly American Rheumatism Association)13), Their mean age at study entry was 35±11 years (mean ± SD), ranging from 14–61 years of age, while the mean disease duration was 7±5 years. Controls were taken from 59 healthy unrelated students of the College of Medical Laboratory Technologists at the Institute for Medical Research, Kuala Lumpur, with no history of any rheumatic diseases and who were ethnically matched. A complete physical and laboratory examination was performed on each patient, while past medical history was taken from previous records. Symptoms noted were; arthritis; mucocutaneous symptoms of malar rash, photosensitivity, oral ulcers and alopecia and renal involvement. Immunological abonormalities noted were antibodies to Sm/RNP, SSA(Ro)/SSB(La) and ds DNA. The age of onset of symptoms were also recorded where patients were divided into 2 groups: below 30 years and at/above 30 years of age. Data were entered into a database while blood samples were obtained for serum and genomic DNA.
1. HLA typing
Genomic DNA was purified from peripheral blood leucocytes using the salting-out method14). DNA typing for “broad” DR groups (DR 1, 2, 3, 4, 5, 6, 7, 8, 9, 10) were determined by PCR while DQA1, DQB1 and DPB1 alleles genotyping was performed by a modified PCR-RFLP, as previously described15,16).
2. Measurement of antiENA and anti ds DNA antibodies
Sera from patients were tested for the presence of autoantibodies to extractable nuclear antigens (including antiSm, antiUIRNP, antiSSA (Ro), antiSSB (La)) and anti ds DNA antibodies using commercial ELISA kits (IMMCO Diagnostics, USA).
3. Statistical analysis
Mhc allele frequency in patients and controls were compared using a 2×2 contigency analysis (×2 test) with Yates correction for each allele or Fishers exact test where appropriate using the EPI-INFO statistical program (Centers for Disease Control, Atlanta, GA), The relative risk was determined by the odds ratio. P value of less than 0.05 was taken to be significant. P corrected (p corr) were determined by multiplying p value with the number of HLA alleles tested. Statistical associations between the clinical and immunological findings and HLA antigens in patients with SLE (antibody positive patients with SLE, antibody negative patients with SLE and controls) were determined by Fishers exact test.
RESULTS
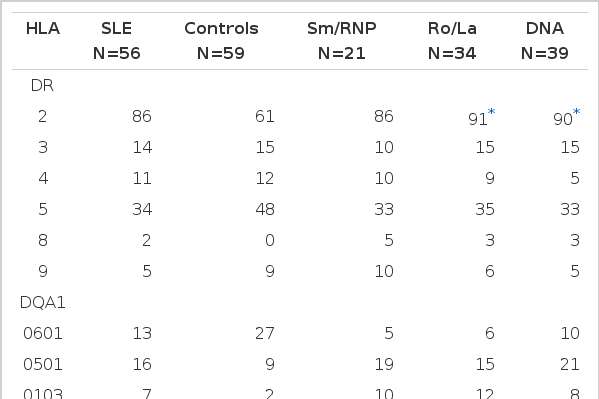
Among our group of 56 Malay patients with SLE, a positive association with SLE was observed for HLA-DR2 (48 of 56, 85.7%, p corr=0.03, rr=3.83) (Table 1). DQB1 *0501 (p corr=0.0036, rr=4.56) and DQB1 *0601 (p corr=0.0048, rr=6.0) (Table 2). There was, however, no DPB specificity linked to SLE disease susceptibility (Table 3). There was a weak decrease of DQA1 *0601 and DQB1 *0503 and *0301 in the patient group with a weak increase of DQB1 *0201 and DPB1 *0901 which did not remain significant after correcting for multiple comparisons made.
1. HLA association with clinical manifestations
Several clinical manifestations were noted and 24 (43%) were found with arthritis, 38(68%) with mucocutaneous symptoms of malar rash, 28 (50%) with photosensitivity, 20 (36%) with oral ulcers, 36 (64%) with alopecia, 38 (68%) with renal involvement. However, immunological abnormalities were seen in several patients: 21 (38%) with antibodies to Sm/RNP, 34(61%) with SSA(Ro)/SSB(La) and 39 (70%) with anti ds DNA antibodies. Twenty patients (36%) were in the younger age group (below 30 years) while thirty-six (64%) were in the older age group (at/above 30 years old).
We analysed the patients who were subgrouped to detemine whether a particular HLA type correlated with the expression of specific clinical manifestations (Table 4). There were positive associations; DR2 with renal involvement (90% vs 78%), DR8 with arthritis (33% vs 3%) when compared to patients without renal involvement and arthritis respectively. However, when comparison was done with healthy controls, there was a positive association of renal involvement with HLA DQB1 *0501 (p corr=0.00084, rr=6.74), arthritis with DQB1 *0501 (p corr=0.00048, rr=9.8), malar rash with DQB1 *0501 (p corr=0.0121, rr=4.41), oral ulcers with DQB1 *0601 (p corr=0.0036, rr=7.2) and alopecia with DQB1 *0501 (p corr=0.00096, rr=6.13). There was no particular HLA specificity with photosensitivity.
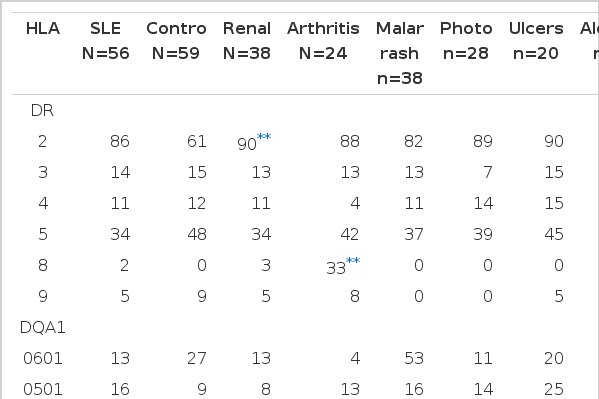
HLA DR, DQ and DP allelelic frequency(%) in controls and SLE patients divided according to their clinical manifestations and age of onset (number of alleles in parentheses)
DQB1 *0501 was also found to be slightly increased (non significant) in the patients with renal involvement compared to those without. However, when similar comparison was made, HLA-DQA1 *0501 was slightly decreased (nonsignificant). DR8 and DQB *0501 was found to be strongly increased in the patients with arthritis compared to those without, but the latter was non significant (p corr=0.03, rr=15.5 and p corr=ns) when corrected for the number of comparisons made. A negative association was found between arthritis and HLA DQB1 *0601 and *0201 though these associations did not remain significant after correction. Similarly, a negative association was noted between malar rash and DQA1 *0103 (nonsignificant association) compared to those without, but when compared to healthy controls no significant association could be observed. HLA DQB1 *0503 was non significantly associated with photosensitivity (p corr=0.18). No other significant associations were observed between genetic factors studied and clinical manifestations seen. We found a weak positive association (nonsignificant) with HLA DPB1 *0301 in those with earlier onset disease compared to those patients with a later onset.
2. HLA association with immunological abnormalities
Twenty-one (37.5%) of 56 patients had antiSm, antiRnp antibodies or both; 34 (60.7%) had antiSSA (Ro) antibodies, antiSSB (La) antibodies or both. Since there is known to be close similarities between the Ro and the La antigens and between the Sm and the RNP antigens17,18), we have chosen to distinguish between antibody responses to these 2 different, non cross-reactive nucleoprotein families. The association between HLA antigens and immunological abnormalities was also analysed to determine if the presence of a particular DR, DQ or DP specificity correlated with autoantibody expression (Table 5). We did not find any correlation of antiSm/RNP and antiRo/La autoantibodies with HLA DR, DQ or DP specificities between antibody positive and antibody negative patients. However, when comparison was made with healthy controls, the prevalence of HLA DQB1 *0601 was found to be significantly increased in antiSm/RNP positive patients (p corr=0.0036, rr=9.82). HLA DQA1 *0601, DQB1 *0501, *0503 and DPB1 *0101 whose frequency in positive patients was 5% vs 27%, 43% vs 17%, 10% vs 39% and 29% vs 9%, respectively, compared with controls but did not reach significance after correction. When the Ro/La positive patients were compared with healthy controls, we found that the prevalence of the HLA DR2(p corr=0.04, rr=6.6) and DQB1*0601 (p corr=0.0098, rr=7.56) were significant in the patient group. There was a slight decrease of HLA DQA1 *0601 and DQB1 *0503 in the patient group when compared to controls but it was not significant after correction was made. Comparing with controls, DR2 was found to be increased in patients with antiRo alone and antiLa alone (p=0.03, and 0.004, respectively, uncorrected) (data not shown). HLA DQB1 *10501 was strongly increased, but DQB1 *0503 was decreased in patients with antiLa alone. When antids-DNA positive patients were compared with healthy controls, we found a strong correlation of HLA DR2, DQB1 *0501, *0601 (p corr=0.04, rr=5.59, p corr=0.0012, rr=6.34 and p corr=0.048, rr=5.4, respectively), with SLE. However, DQB1 *0503 and *0301 were found in excess in the control group but did not reach statistically significant value after correction for multiple comparisons made.
DISCUSSION
Since genetic factors play a role in the predisposition to the disease arising from the high concordance rate for SLE in identical twins19) and the tendency for familial aggregation in SLE20) and that the mhc has been linked to this, it is of interest whether clinical manifestations and/or the expression of autoantibodies are associated with the HLA antigens. Here, in this present study, we have set to examine whether the HLA complex genes (HLA class II antigens) are implicated in the predisposition to SLE and to determine whether these loci correlate with the presence of clinical manifestations and immunological abnormalities.
We found a significantly increased frequency of DR2, DQB1 *0501 and DQB1 *0601 in patients with SLE compared to normal controls. However, DQA1 *0601 and DQB1 *0503 and *0301 were found to be nonsignificantly decreased while DQB1 *0201 and DPB1 *0901 were nonsignificantly increased in SLE. Our finding of the association of the DR2 with SLE is in agreement with that found by others21–24), while DR3 antigens were found to be associated with the Caucasian population6) and Black SLE patients7). However, others did not observe any HLA association with SLE25,26).
The association of particular HLA specificities and clinical and immunological expression and/or severity of disease have been reported before11,27–30) with inconsistent results. A study on HLA DR and DQ alleles showed an association SLE with DR3 and DQw2.1 but no DR or DQ gene associations could be found with clinical manifestations in Caucasians28,29). Few studies have examined the associations of DP antigens and disease, probably because of the difficulties in typing by cellular methods. HLA DP antigens are expressed in lower densities on the cell surface than DR and DQ and, therefore, the last of the functional HLA class II loci to be described. They were initially defined at the cellular level by primed lymphocyte typing31).
In our study, we found some positive and negative association between HLA antigens and manifestations of clinical and immunological features. There was a strong positive association of DR2 and DQB1 *0501 in patients with renal involvement with a weak decrease of DQA1 *0501. An increase frequency of DR2 was reported in patients with lupus nephritis in Caucasian (American), Blacks, Asians and Pacific Islanders32). However, a significant decrease of DR2 was seen in patients with renal involvement21). A weak association of DR5 with renal involvement, almost due to the linkage of DR5 to DRw52b haplotypes, was also found28). In another study, no association of DR2 or 3 or any other DR or DQ specificity with renal involvement was noted29).
DR8 was strongly associated with our group of patients with arthritis, while DQB1 *0601 with oral ulcer and DQB1 *0501 with malar rash, but no association with photosensitivity. Our observation of an association of DQB1 *0501 with alopecia was different from that found by Hong et al.21) where his association was with DR9. We found that there were non significant associations of DQB1 *0501, 0601 and 0201 with arthritis, DQA1 *0103 with malar rash, DQB1 *0503 with photosensitivity. Therefore, it is probable that the majority of all these associations resulted from coincidence. Positive associations of photosensitivity with DRB1 *0405 and/or DQB1 *0401, malar rash with DPB1 *0201, oral ulcers with DRB1 *0901 and alopecia with DPA1 *0201 were noted but no association with arthritis was observed in another study33).
We did not observe any DR or DQ association with early or late disease onset. Only HLA DPB1 *0301 was found to be non significantly associated with patients in the earlier onset disease in our group of patients. An increased frequency of DR3 in late onset SLE34) was not observed by Davies et al.35) who instead found the situation to be reversed, with DQA1 *0501, DR3 frequencies elevated in those patients with onset before the age of 30 years compared to those above 30 years of age and DR5, which is in linkage disequilibrium with DQA1 *0501 was also found to be non significantly elevated in those with earlier onset SLE. In his group of patients, DR4 was more common in those patients with later onset SLE but this was statistically nonsignificant. These differences in the immunogenetic profiles of early and late onset SLE, with the classical markers particularly prevalent in those with onset before the age of 30 years, may help explain some of the differences between early and late onset SLE36).
Associations of the presence of SSA (Ro) and SSB (La) antibodies with DR and DQ antigens have been reported11,37,38). We found a significant association of the DR2 with the Ro/La antibody complex with a non significant decrease of DQA1 *0601 and DQB1 *0503. DR2 was also found to be associated with Ro/La antibodies previously21) but was negated by others28,29). Others have found HLA DR3 to be associated with antiRo antibodies in SLE10–12,39). As for the DQ genes, both DQw2 and DR3 were found to be increased in the antiRo/La subgroup, while in the antiSm/RNP subgroup DQw2 and DR4 were found to be increased40). Thus, it is apparent that both DR and DQ antigens may play a role in antibody production in SLE but does not necessarily mean that a cooperation has to exist between these DR and DQ alleles. Very few patients in his study with antiRo/La or antiSm/RNP antibodies had DR3 or DR4, respectively, with DQw2. The various genetic associations in different races may be partially due to their different genetic background which could contribute to the pathogenesis and clinical course of disease.
We did not observe any DP specificity in patients positive for antiRo/La antibodies. Two groups have reported the results for an association between Ro and La antibodies and DPB1 alleles41,42). A slightly higher frequency of *0101 in anti Ro/La negative patients, which was nonsignificant, was noticed41). However, a non significant increase of *0101 in antiRo positive patients with or without La antibodies, as compared with patients without these antibodies and with healthy controls, was observed in another study42).
Though several studies have noticed the association of DR2 or DR4 with either antiSm or RNP antibodies39,43). We and two other groups of investigators10,11) could not confirm this. Our analysis of correlation between HLA and antiSm/RNP antibodies revealed the association of DQB1 *0601 in our study. However, there was a nonsignificant increase of DQA1 *0601, DQB1 *0501 and *0503 which could have occurred by chance. There was no HLA association with antiSm alone but we found a significant association of DQB1 *0601 with antiRNP alone. AntiSm or antiRNP or both were found to be associated with DR4 and somewhat more strongly with the presence of DR4 or DR1 or both30). The low frequency of DR3 (known to be closely associated with antiRo/La) noted by Alvarellos et al.12) indicated that the expression of antiSm/RNP is genetically different from that of antiRo/La. The molecular structures of the related antigen Ro and La differ from the structure of the related antigen Sm and RNP and that antiLa almost always occurs together with antiRo, whereas antiSm is very often accompanied by antiRNP10,11). DR5 was found to be related to the absence of these autoantibodies and thus appeared to protect against their expression. A positive association between antiRNP and DQB1 *0410 was found33).
The role of the DPB locus has also been studied in antiSm and RNP autoantibody production. DPB1 *0401 has been reported as being associated with the presence of antibodies to RNP44) in connective tissue disease patients. We found a weak (nonsignificant) association of DPB1 *0901 with antiSm/RNP antibodies. AntiRNP was frequently found in those with DPB *0301 (statistically insignificant due to the small number of SLE patients in this group)42) while another study found an increased frequency of DPB1 *0301 and *1401 in SLE patients with antiSm/RNP41).
A strong correlation of DR3 with antibodies to dsDNA was earlier reported45). However, we observed a strong association of DR2, DQB1 *0501 and *0601 in our SLE patients. Others reported high levels of anti DNA antibodies associated with DR2 and possibly DQ111,12) while Schur et al4) noted DR7 to be increased in antids DNA positive patients. There was a trend toward an increase of DR2, DQB1 *0501 and *0601 in patients with antibodies to DNA27. These findings were in contrast with others29,39). This could be due to ethnic/racial differences and clinical heterogeneity of the patients in the different study groups.
Our data suggest that a marked clinical and immunogenetic heterogeneity of SLE have been demonstrated and that these clinical and immunological expressions may be controlled by the genes within the major histocompatibility complex.
Acknowledgements
This work was partially supported by the IRPA grant 06-05-01-0121. We would like to thank the Director of the Institute for Medical Research for permission to publish this article.