|
 |
| Korean J Intern Med > Volume 12(1); 1997 > Article |
|
Abstract
Objectives
The relationship between HCV genotype and the development of more serious liver disease has not been clearly established. This study was to investigate the distribution pattern of HCV genotypes in Korea and their relationship to the viremic level and to progression of chronic liver disease.
Methods
Study population was 217 patients with type C chronic liver disease. They were divided into 4 groups: 83 patients with near-normal ALT (group 1), 64 patients with elevated ALT (group 2), 20 patients with decompensated liver cirrhosis (group 3) and 50 patients with hepatocellular carcinoma (group 4). HCV genotypes were determined by reverse transcription polymerase chain reaction (RT-PCR) using mixed primer sets, and then the fidelity of genotyping was confirmed by cloning and sequencing. HCV RNA concentration was measured by quantitative competitive RT-PCR for 23 patients in group 2.
Results
The genotypes could be determined in 166 (76%) out of 217 patients. Type 1b and type 2a were predominantly occurring over the other types in somewhat similar frequency (45% and 51%, respectively). The genotype distribution of type 1b and 2a among four different groups showed 42% and 54% in group 1, 49% and 45% in group 2, 53% and 47% in group 3 and 41% and 57% in group 4; thus there was no significant difference in genotype distribution among 4 different disease groups. However, the viremia levels in patients with genotype 1b infection were significantly higher than those with genotype 2a.
Chronic hepatitis C is typically a slowly progressive, asymptomatic disorder that is carcinoma (HCC) in some patients1). An astonishingly high proportion of individuals acutely infected with hepatitis C virus (HCV) become chronically infected (approximately 80%), and more than 20% of these individuals will then develop liver cirrhosis and eventually HCC2). Although hepatitis B virus is a major etiologic agent in chronic liver diseases and HCC in Korea, it has been revealed that HCV also plays an important role3,4).
One of the most important characteristics of HCV is genetic heterogeneity of its genome. Various HCV isolates can be divided into genetically distinct groups as a result of accumulation of mutations during the evolution of these viruses5ŌĆō7). A total of nine such major genetic groups and at least 30 subgroups have been identified to date on analysis of complete or partial HCV genomic sequences8,9). Although many genotypes are widely distributed around the world, there are clearly distinct differences in their regional distribution8,10). The predominant HCV genotype was reported to be type 1b in Korea11). However, the fidelity of genotyping has not been checked by DNA sequencing.
Besides epidemiological significance, HCV genotypes have also important diagnostic and clinical implications12,13). Recently, a number of studies have shown their influence on the progression of liver disease and on the outcome of interferon therapy in HCV-infected patients. Genotype 1b is reported to be more prevalent in patients with cirrhosis and HCC than in patients with chronic hepatitis14ŌĆō16). However, the findings in these studies were not considered against a background of predominant type 1 infection in the countries where these studies were performed. Thus, the relationship between HCV genotypes and the development of more serious liver disease has not been clearly established yet. It now seems to be fairly well documented that patients infected with genotype 1b respond less well to interferon therapy than those infected with genotypes 2a or 2b. HCV types might show different rates of replication and thus different levels of viremia17ŌĆō19). Therefore, the lower response to interferon in type 1b may be due to higher viremia, and results in poor clinical outcome.
The present study was to investigate the diversity of HCV genotypes in Korea and its relationship to the viremic level and to the progression of chronic liver disease.
We consecutively enrolled 217 patients who tested positive either for anti-HCV or for serum HCV RNA by RT-PCR. Those who simultaneously had HBsAg in their sera were excluded. The patients consisted of 200 anti-HCV-positive patients and 17 HCV RNA-positive patients out of 89 anti-HCV-negative patients with chronic liver diseases. Among 200 patients who tested positive for anti-HCV, the positivity of HCV RNA was 75% (149/200). The mean age of 217 patients was 56┬▒ 12 years (range, 13ŌĆō78 years) with a male to female ratio of 1.8(139/78). They were divided into four groups; 1) group 1 consisted of 83 patients with persistently normal or near normal ALT levels. Near normal ALT level was defined as occasional ALT fluctuations within 5 IU above the upper limit of normal (40 IU/L); 2) group 2 consisted of 64 patients who presented with elevated ALT but were asymptomatic and did not show any sign of portal hypertension. Although liver biopsy was not performed in most of these patients, this group was supposed to reflect the patients with chronic hepatitis or compensated chronic liver disease; 3) group 3 consisted of 20 patients who presented with any clinical sign of portal hypertension such as ascites, variceal bleeding or hepatic encephalopathy. This group of patients reflected those with decompensated liver cirrhosis; 4) group 4 consisted of 50 patients with HCC. The diagnosis of HCC was made as previously described3). The mean ages of group 1, 2, 3 and 4 were 52, 52, 59 and 64 years, respectively. The mean age of group 3 or group 4 was significantly higher than those of group 1 or group 2.
The sera that had been collected on the first visit were tested for HBsAg (AUSRIA-II, Abbott Laboratories, Chicago IL) and anti-HCV (HCV-EIA, Abbott Laboratories, North Chicago, IL) using commercially available kits. Aliquots of sera were stored at ŌłÆ70┬░C for reverse transcription polymerase chain reaction (RT-PCR) to detect HCV RNA.
HCV RNA detection and genotyping was performed by nested RT-PCR using type-specific primers. Extraction of HCV RNA and reverse transcription was performed according to Romeo et al20). PCR of HCV cDNA was performed by Chayama et al21) with some modifications. The modifications were as follows; amplification of the cDNA was performed by nested PCR instead of one step method, in which the first round PCR was performed with universal primers (#13, #14) and the second round PCR was performed with mixed primer sets (No. 6 and No. 7)21). No. 6 was equimolar mixtures of anti-sense primer #14 and sense primers #121, #61, #59, #Tr7 and #111, each concentration of which was 10 umol/L, respectively. No. 7 was equimolar mixtures of antisense primer #14 and sense primers #57, #117, #113, #Tr7 and #111. Each of the amplifications consisted of denaturation at 94┬░C for 3 min and extention at 72┬░C for 7 min and then 35 cycles of denaturation at 94┬░C for 30 sec, annealing at 57┬░C for 30 sec and extention at 72┬░C for 30 sec, and finally extention at 72┬░C for 5 min, respectively. Each genotype was determined by its characteristic DNA size21).
Genotyping fidelity was confirmed by two ways. Firstly, 50 random serum samples, the genotypes of which had been tested in our laboratory, were sent to another laboratory in Japan (Osaka Red Cross Blood Center). Genotypes were determined there using type-specific primers derived from HCV core region, different from ours which were derived from NS5 region. Secondly, the genotypes of 20 random samples were determined by cloning and sequencing of NS5 region.
Serum samples from 23 patients which consisted of 14 type 1b and 9 type 2a, randomly selected in group 2, were measured for HCV RNA concentrations. Quantitation was performed by competitive RT-PCR using nested primers. The outer primers were JR12 and JR19, and the inner primers were JR13 and JR1420). The size of internal control HCV RNA was larger than native HCV RNA by 71 base pairs. After the extraction of RNA from 60uL serum, 1:10 volumes of RNA were mixed with serial dilutions of internal control RNA. Samples were amplified in the same way as that for genotype determination20).
Only viremic patients in whom HCV genotype could be detected were considered for statistical analysis. X2 test was used as appropriate for analysis of genotype distribution in different groups. Comparison of HCV RNA concentrations (logarithm of copies/mL) was performed by Wilcoxon rank sum test.
Among 50 serum samples the genotypes of which were tested again in Osaka Red Cross Hospital, the concordance rate was at least 90% (45/50). Among 20 serum samples, the genotypes of which were confirmed by cloning and sequencing, the concordance rate was 90% (18/20).
HCV RNA was detected and, therefore, the genotypes could be determined in 166(76%) out of 217 patients. The distribution of genotypes in these 166 patients is shown in Table 1. Type 1b and type 2a were predominant (45% and 51%, respectively) over the rest of the genotypes (1a, 2b, 3), the percentage of which was less than 3% in total. There were no significant differences in male-to-female ratio (1.3 vs 2.1) and mean age (57.6 vs 54.3 years) between patients with type 1b infection and those with type 2a infection.
The positive rates of HCV RNA were 71% (59/83) in group 1, 83% (53/64) in group 2, 85% (17/20) in group 3 and 74% (37/50) in group 4, and they were not statistically different among different groups (p=0.3). In each group, both type 1b and type 2a were predominant over the rest (Table 1). Frequencies of major genotypes (1b and 2a) were not significantly different among four groups (p=0.7), although frequency of type 1b tended to increase as the disease progressed. Type 1a which is most prevalent in the United States was found in only two (0.8%) patients.
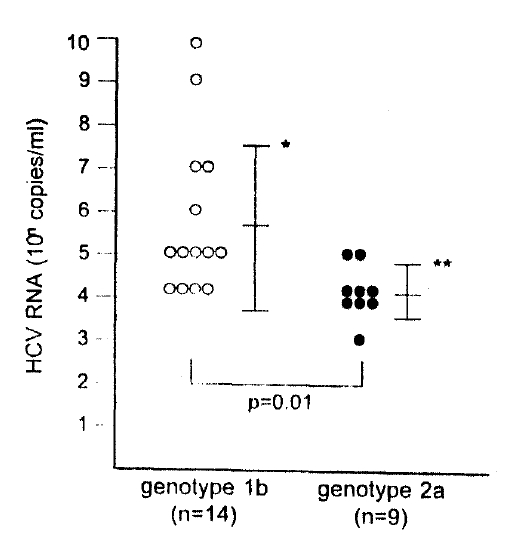
HCV RNA concentration (logarithm of copies/mL) was 5.7+1.9 (mean ┬▒ SD) in patients with genotype 1b infection and 4.1 ┬▒0.6 in patients with genotype 2a infection. The difference was significantly different (p=0.01) (Fig. 1).
The analysis of the diversity of HCV genotype in a country is important for understanding the molecular basis of the route of transmission of HCV as well as viral virulence and interferon resistance. RT-PCR is the mainstay of HCV genotype determination, followed by several different methods; (1) a second round of PCR amplification with type-specific primers; (2) hybridization with type-specific probes; (3) digestion with different restriction enzymes to determine a specific restriction fragment length polymorphism (RFLP)8); or (4) analysing sequences directly or after cloning. However, methods that do not include sequence analysis are not definitive.
Our present study disclosed that, among Korean patients with type C chronic liver disease, the predominant HCV genotype was type 1b and type 2a (45% vs 51%) and the prevalence rates of the two genotypes were similar. In contrast, the previous study in Korea which was not confirmed by sequence analysis showed that genotype 1b was predominant (75%) over 2a (25%). This discrepancy might result from various technical factors previously described or it can be attributed to clinical factors. In the present study, the technical factor for the potentially incorrect genotyping was evaluated with sequence analysis in 20 samples, which gave the concordance rate of 90%. Therefore, the fidelity of this study might be considered as acceptable. With regard to clinical bias, the number of enrolled patients and the method of patient selection can affect the result. For example, the genotype distribution can vary significantly in different geographic areas of the same country, as has been observed in China22). Furthermore, the observed geographical distribution of HCV genotypes could vary depending on which group of individuals was studied12,23,24). The genotype distribution has been found to be different in patients with chronic liver disease compared to blood donors23,24). In addition, the genotype distribution has differed in intravenous drug abusers compared to other groups of HCV-infected individuals12). The result of the present study is considered to more accurately represent the genotype distribution than the previous one because of the inclusion of a larger number of patients and the inclusion of patients with variable chronic liver diseases. This distribution pattern in Korea is unique among Asian countries, because genotype 1b is more predominant in most Asian countries than 2a8). The proportion of genotype 1a and genotype 2a has been reported to be 68% and 24%, respectively, in China, 74% and 24%, respectively, in Japan and 65% and 30%, respectively, in Taiwan.
In the present study, there was no significant difference in the distribution of HCV genotypes at different stages of chronic liver disease, suggesting that progression of type C chronic liver disease to decompensated liver cirrhosis or development of HCC is not strongly associated with HCV genotypes. These findings are consistent with previous reports from Japan24ŌĆō26). In contrast, Pozzato and co-workers14) in Italy found that infection with HCV isolates of genotype 1b was associated with more severe liver disease than infection with HCV isolates of other genotypes. There are several possible explanations for this discrepancy. Firstly, the background genotype distribution pattern might affect the result; the role of the genotype 1b in advanced liver disease might be exaggerated where the genotype 1b was predominant. Secondly, small number of enrolled patients or confounding factors such as age and duration of viral infection might cause clinical bias. In fact, in the study of Pozzato et al14), the mean age of patients infected with genotype 1b was significantly higher than that of patients infected with other genotypes, and the number of patients was not large enough. In our study, age and sex differences were not observed between genotype 1b and 2a, although the duration of viral infection could not be evaluated because most of the patients had sporadic community-acquired infection. Prospective long-term follow-up studies are also warranted to clarify the association of specific genotype with more likelihood of progression to advanced liver disease in various geographical areas, especially in areas where genotypes other than 1b are as prevalent as type 1b.
HCV RNA concentrations were significantly lower in patients with type 2a than in those with type 1b. The result is consistent with some previous reports17ŌĆō19) but not with others which did not observe such a difference in the level of viremia26ŌĆō28). This discrepancy might result from the difference in the study design or the variability of viremia level in a patient over time. Irrespective of viremia level, it seems that genotype is less likely to affect the long-term clinical outcome because there was no significant difference in the overall clinical outcome among HCV genotypes, as shown in this study. Longitudinal study to determine HCV RNA levels in sequential serum samples obtained from a large number of patients may be required to make clear this point.
There is substantial evidence that the genotype status of HCV-infected patients is important in determining the outcome of interferon therapy. Multivariate analysis of factors that might predict the outcome of interferon therapy in patients chronically infected with HCV demonstrated that genotype status, liver histology and pretreatment level of viremia are important predictive factors 27,29,30). Among them, genotype status most efficiently contributed to the treatment outcome30); infection with type 2a or 2b predicted a more favorable response to interferon therapy than infection with type 1b (78% vs. 9%)30). The difference in therapeutic response according to genotype, together with the finding that the viremic level can be different among different genotypes, suggests that there are particular groups (eg. genotype 2a) of patients who might get maximal benefit from antiviral therapy.
In summary, we found that type 2a HCV infection is as prevalent as 1b in Korea. Although there is a difference in viremia level among patients with different genotype infection, it seems that they have similar risk for progression to advanced diseases. On the basis of differences in response to antiviral therapy among genotypes and the higher prevalence of genotype 2a in Korea, it is warranted to see if the response rates of chronic viral hepatitis C to interferon are higher than those in other countries, although it still remains to be seen whether there will be a greater increase in survival time in treated than in untreated patients with the passage of a few decades.
Notes
This work was supported by grant No. 02-94-002 from the Seoul National University Hospital Research Fund, and by a research grant from Liver Research Foundation of Korea (1995), and grant ┬Ę in ┬Ę aid for international scientific program special cancer research from the Ministry of Education, Science and Culture, Japan
Fig.┬Ā1.
Comparison of HCV RNA concentration in serum between HCV genotypes. Mean serum HCV RNA concentrations in cases with genotype 1b (mean┬▒SD: 5.7┬▒1.9 log copies/ml) significantly exceeded that in cases with genotype 2a (mean┬▒SD: 4.1 ┬▒0.6 log copies/ml) (p=0.01).
Table┬Ā1.
Distribution of HCV Genotypes in Different Patient Groups
Group 1: patients with near-normal ALT; Group 2: patients with elevated ALT; Group 3: patients with decompensated liver cirrhosis; Group 4: patients with hepatocellular carcinoma. HCV genotype was classified according to Simmonds et al9).
REFERENCES
1. Alter HJ. Transfusion-associated non-A, non-B he patitis: The first decade. In: Zuckerman AJ, ed. Vira hepatitis and liver disease. 537. New York: Alan R Liss, 1988.
2. Alter HJ, Seeff LB. Transfusion-associated hepatitis. In: Zuckerman AJ, Thomas HC, eds. Viral hepatitis scientific basis and clinical management. 467. Edinburgh: Churchill Livingstone, 1993.
3. Lee H-S, Han CJ, Kim CY. Predominant etiologic association of hepatitis C virus with hepatocellula carcinoma compared with hepatitis B virus in elderly patients in a hepatitis B-endemic area. Cance 1993;72:2564.

4. Kim CY, Lee H-S, Han CJ. Relative etiologic role of hepatitis B virus and hepatitis C virus in chronic liver diseases and hepatocellular carcinoma among age-specific groups in Korea: the possible presence of non-B, non-C agents. Seoul J Med 1993;34ŌĆō27.
5. Houghton M, Weiner A, Han J. Molecular biology of the hepatitis C viruses: implications for diagnosis development and control of viral disease. Hepatology 1991;14:381.


6. Okamoto H, Kurai K, Okada SI. Full-length sequence of a hepatitis C virus genome having poo homology to reported isolates: comparative study o four distinct genotypes. Virology 1992;188ŌĆō331.
7. Bukh J, Purcell RH, Miller RH. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci USA 1993;90:8234.



8. Bukh J, Miller RH, Purcell RH. Genetic heterogeneity of hepatitis C virus: quasispecies and geno types. Sem Liver Dis 1995;15:41.

9. Simmonds P, Holmes EC, Cha T-A. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol 1993;74:2391.


10. Dusheiko G, Schmilovitz-Weiss H, Brown D, McOmish F, Yap P-L, Sherlok S, Mclntyre N, Simmonds P. Hepatitis C virus genotypes: an investigation of type-specific differences in geographic origin and disease. Hepatology 1994;19:13.


11. Yoon SK, Park YM, Chung KW, Kim BS, Kim CJ, Kim WY, Chang SK, Cho MJ. Molecular typing of hepatitis C virus genome from sera and liver tissues of patients with anti-HCV positive chronic liver disease. Kor J Int Med 1993;8:66.



12. Brechot C. Hepatitis C virus genetic variability: clinical implications. Am J Gastroenterol 1994;89:S41.

13. Bukh J, Miller RH. Diagnostic and clinical implications of the different genotypes of hepatitis C virus Hepatology 1994;20:256.
14. Pozzato G, Kaneko S, Moretti M, Croce LS, Franzin F, Unoura M, Bercich L, Tiribelli C, Crovatto M, Santini G, Kobayashi K. Different genotypes of hepatitis C virus are associated with different se verity of chronic liver disease. J Med Virol 1994;43:291.


15. Qu D, Li J-S, Vitvitski L, Mechai S, Berby F, Tong S-P, Bailly F, Wang Q-S, Martin J-L, Trepo C. Hepatitis C virus genotypes in France: comparison of clinical features of patients infected with HCV type I and type II. J Hepatol 1994;21:70.


16. Silini E, Bono F, Cividini A, Cerino A, Bruno S, Rossi S, Belloni G, Brugnetti B, Civardi E, Salvaneschi L, Mondelli MU. Differential distribution of hepatitis C virus genotypes in patients with and without liver function abnormalities. Hepatology 1995;21:285.


17. Kobayashi Y, Watanabe S, Konishi M. Quantitation and typing of serum hepatitis C virus RNA in patients with chronic hepatitis C treated with inter feron-╬▓. Hepatology 1993;18:1319.


18. Sherman K, OŌĆÖBrien J, Gutierrez A. Quantitative evaluation of hepatitis C virus RNA in patients with concurrent human immunodeficiency virus infections. J Clin Microbiol 1993;31:2679.



19. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987;162:156.


20. Romeo JM, Ulrich PP, Busch MP, Vyas GN. Analysis of hepatitis C virus RNA prevalence and surro gate markers of infection among seropositive vo luntary blood donors. Hepatology 1993;17:188.


21. Chayama K, Tsubota A, Arase Y, Saitoh S, Koida I, Ikeda K, Matsumoto T, Kobayashi M, Iwasaki S, Koyama S, Morinaga T, Kumada H. Genotypic subtyping of hepatitis C virus. J Gastroenterol He patol 1993;8:150.

22. Wang Y, Okamoto H, Tsuda F. Prevalence, genotypes, and an isolate (HC-C2) of hepatitis C virus in Chinese patients with liver disease. J Med Viro 1993;40:254.

23. Okamoto H, Sugiyama Y, Okada S. Typing of hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol 1992;73:673.


24. Ichimura H, Tamura I, Kurimura O, Koda T, Mizui M, Tsuchie H, Kurimura T. Hepatitis C virus genotypes, reactivity to recombinant immunoblot assay 2 antigens and liver disease. J Med Virol 1994;43:212.


25. Yamada M, Kakumu S, Yoshioka K, Higashi Y, Tanaka K, Ishikawa T, Takayanagi M. Hepatitis C virus genotypes are not responsible for development of serious liver disease. Dig Dis Sci 1994;39:234.


26. Mita E, Hayashi N, Kanazawa Y, Hagiwara H, Ueda K, Kasahara A, Fusamoto H, Kamada T. Hepatitis C virus genotype and RNA titer in the progression of type C chronic liver disease. J Hepatol 1994;21:468.


27. Tsubota A, Chayama K, Ikeda K, Yasuji A, Koida I, Saitoh S, Hashimoto M, Iwasaki S, Kobayashi M, Hiromitsu K. Factors predictive of response to inter-feron-╬▒ therapy in hepatitis C virus infection. Hepatology 1994;19:1088.


28. Takase S, Takada N, Sawada M. Effects of interferon treatment for chronic type C liver diseases with different hepatitis C virus genotypes: a cooperative study in three hospitals. Int Hepatol Commun 1993;1:321.




 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print