|
 |
| Korean J Intern Med > Volume 27(4); 2012 > Article |
|
Abstract
Background/Aims
The relationship between Runt-related transcription factor 3 (RUNX3) gene inactivation and various solid tumors has been reported; however, little information is available about RUNX3 in thyroid cancers.
Methods
We evaluated the DNA methylation of RUNX3 in 13 papillary thyroid cancer tissues and four thyroid cancer cell lines. Additionally, using reverse transcriptase-polymerase chain reaction, we analyzed RUNX3 gene expression in several thyroid cancer cell lines after treating with the demethylating agent 5-aza-2'-deoxycytidine (DAC).
Runt-related transcription factor 3 (RUNX3) has strong tumor suppressor activity, which is involved in the regulation of epithelial proliferation and apoptosis [1,2]. Inactivation of the RUNX3 gene has been reported in many human solid tumors. The RUNX family consists of three members: RUNX1 (PEBP2Ab/CBFA2/AML1), RUNX2 (PEBP2Aa/CBFA1/AML3), and RUNX3 (PEBP2Ac/CBFA3/AML2). RUNX1, located on chromosome 21q22.3, is related to hematopoiesis, and its mutation has been reported in human acute leukemia and myelodysplastic syndrome. RUNX2, located on chromosome 6q21, has been linked to bone formation. RUNX3, located on chromosome 1p36, plays an important role in the development of cancer [3-5].
RUNX3 gene inactivation has been reported in gastric cancer, colorectal cancer, lung cancer, pancreatic ca ncer, bladder cancer, breast cancer, and hepatocellular carcinoma [6-15]. However, little information is available about the role of RUNX3 in thyroid cancers. To determine whether methylation of RUNX3 is related to carcinogenesis of the thyroid, we evaluated DNA methylation of RUNX3 in 13 papillary thyroid cancer tissues and four thyroid cancer cell lines. RUNX3 gene expression in some thyroid cancer cell lines after 5-aza-2'-deoxycytidine (DAC) treatment was also analyzed using reverse transcriptase-polymerase chain reaction (RT-PCR).
Human thyroid tissues collected during surgery were immediately frozen in liquid nitrogen. We examined 13 tumor tissue samples (12 papillary thyroid carcinomas [PTCs] and one Hurthle cell adenoma), and five normal thyroid tissues, which consisted of normal tissues adjacent to the thyroid tumors. All tumors were classified according to the criteria of the World Health Organization Committee. One human follicular cancer cell line (WRO), two human papillary cancer cell lines (TPC1 and K1), and one anaplastic cancer cell line (HTH74) were also investigated. These cell lines were kindly provided by Dr. Adel K. El-Naggar (M.D. Anderson Cancer Center, University of Texas, Houston, TX, USA).
DNA was extracted using standard phenol/chloroform methods. Genomic DNA (2 µg) was denatured in 2 M NaOH for 10 minutes at 37℃. The denatured DNA was diluted in 30 µL freshly prepared solution of 10 mM hydroquinone (Sigma-Aldrich, St. Louis, MO, USA) and 520 µL freshly prepared solution of 3 M sodium bisulfite (Fisher Scientific, Hampton, NH, USA) at pH 5.0, and incubated for 16 hours at 50℃. After incubation, the DNA sample was desalted through a column (Wizard DNA Clean-Up System, Promega, Madison, WI, USA). The DNA was eluted using 50 µL warm water and treated with 5.5 µL 3 M NaOH for 5 minutes at room temperature, and precipitated with ethanol with glycogen as a carrier. The bisulfite-modified genomic DNA was resuspended in 20 µL H2O and stored at -20℃ until use.
The sodium bisulfite-modified DNA was amplified by PCR. The primers used to amplify RUNX3 were as follows: 5'-TTT GGA GAT ATT TGG GTT TT-3' (sense) and 5'-CCC ATT TAA TAT ACA CAC AAC TAA-3' (antisense). The PCR conditions were as follows: 35 cycles of denaturation at 94℃ for 30 seconds, annealing at 60℃ for 30 seconds, and finally a extension at 72℃ for 30 seconds. PCR products were digested with HpyCH4IV (New England BioLabs, Ipswich, MA, USA). The enzyme-treated DNA products were separated by electrophoresis on 5% PAGE gels. DNA from the blood of normal healthy individuals was used as a negative control. SssI methylase-treated (New England Biolabs) normal lymphocyte DNA was used as a positive control.
Thyroid cancer cell lines were cultured in RPMI1640 supplemented with 10% fetal bovine serum. Two cell lines (WRO and TPC1) were treated with a final concentration of 0.5, 1, and 5.0 µM DAC (Sigma) for 3 days, and/or 300 nM trichostatin A (TSA; Sigma) on the third day. Cells were harvested on the fifth day of treatment and subjected to RNA extraction.
Total RNA was purified from the cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed to single-stranded complementary DNA using random hexamers and MMLV reverse transcriptase. The primers used to amplify RUNX3 were as follows: 5'-ACG CCT ACG TCA TCC TGA AA-3' (sense) and 5'-ATG CCA CAC CTC CTT TCT TA-3' (antisense). Glyceraldehyde 3-phosphate dehydrogenase was used as an internal reference gene. Amplified products were resolved by agarose gel electrophoresis and visualized with ethidium bromide.
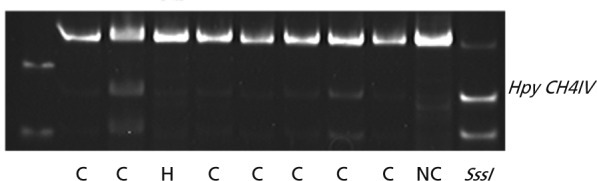
COBRA was performed to analyze the methylation status of the RUNX3 CpG islands in human thyroid cancer cell lines and thyroid cancer tissues. Almost all thyroid cancer cell lines showed increased methylation of RUNX3 (Fig. 1). Among the thyroid cancer cell lines studied, WRO and TPC1 had extremely hypermethylated RUNX3, at over 90% (Table 1). RUNX3 was hypermethylated in 10 of the 12 papillary thyroid cancer tissues and was not methylated in five normal thyroid tissues. One Hurthle cell adenoma tissue was not hypermethylated (Fig. 2).
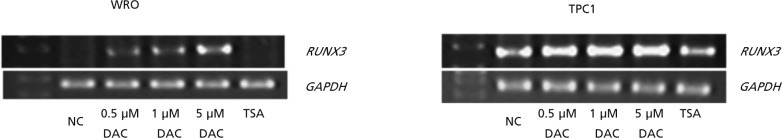
Two human thyroid cancer cell lines (WRO and TPC1) were treated with the demethylating agent DAC and histone-deacetylating agent TSA in an attempt to reactivate RUNX3. DAC treatment resulted in a dose dependent increase in expression of RUNX3 (Fig. 3).
Recently, many reports have described the association of RUNX3 gene inactivation with different solid cancers. However, little information is available on the relationship between RUNX3 and thyroid cancer. In this study, we analyzed the methylation status of RUNX3 in thyroid cancer and showed that RUNX3 was hypermethylated in various thyroid cancer cell lines and PTC tissues.
Promoter hypermethylation is considered to be a main mechanism for gene inactivation. Hypermethylation of CpG islands located in the promoter regions of tumor suppressor genes is associated with cell pathway disruption and transcriptional inactivation. CpG island hypermethylation has been reported in stomach cancer, hepatocellular carcinoma, lung cancer, breast cancer, and pancreatic cancer. RUNX3 is silenced by hypermethylation of CpG islands in the promoter region [16,17].
The exact tumor suppressive action of RUNX3 is not clear. RUNX3 is an integral component of TGF-β that induces cell signal pathways. The gastric epithelium of RUNX3 knock-out mice show reduced apoptosis and decreased sensitivity of the RUNX3 gene-associated TGF-β signaling pathway [18]. TGF-β regulates the G1 phase of the cell cycle, and reduced TGF-β sensitivity induces growth arrest associated with apoptosis [19,20].
In this study, RUNX3 hypermethylation was present in papillary thyroid cancer cell lines, follicular thyroid cancer cell lines and anaplastic thyroid cancer cell lines. We evaluated RUNX3 only in papillary thyroid cancer tissues, which showed a high methylation rate. Although a similar evaluation of RUNX3 in other thyroid cancer types is needed, our results suggest that hypermethylation of RUNX3 may be associated with thyroid cancer development, at least with PTC.
In conclusion, RUNX3 is associated with thyroid cancer and RUNX3 methylation is a potentially useful diagnostic marker for PTC.
References
1. Coffman JA. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol Int 2003;27:315–324PMID : 12788047.


2. Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer 2005;5:376–387PMID : 15864279.


3. Bae SC, Ogawa E, Maruyama M, et al. PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol 1994;14:3242–3252PMID : 8164679.



4. Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics 1994;23:425–432PMID : 7835892.


5. Avraham KB, Levanon D, Negreanu V, et al. Mapping of the mouse homolog of the human runt domain gene, AML2, to the distal region of mouse chromosome 4. Genomics 1995;25:603–605PMID : 7790005.


6. Li QL, Ito K, Sakakura C, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 2002;109:113–124PMID : 11955451.


7. Oshimo Y, Oue N, Mitani Y, et al. Frequent loss of RUNX3 expression by promoter hypermethylation in gastric carcinoma. Pathobiology 2004;71:137–143PMID : 15051926.


8. Goel A, Arnold CN, Tassone P, et al. Epigenetic inactivation of RUNX3 in microsatellite unstable sporadic colon cancers. Int J Cancer 2004;112:754–759PMID : 15386381.


9. Araki K, Osaki M, Nagahama Y, et al. Expression of RUNX3 protein in human lung adenocarcinoma: implications for tumor progression and prognosis. Cancer Sci 2005;96:227–231PMID : 15819721.


10. Li QL, Kim HR, Kim WJ, et al. Transcriptional silencing of the RUNX3 gene by CpG hypermethylation is associated with lung cancer. Biochem Biophys Res Commun 2004;314:223–228PMID : 14715269.


11. Li J, Kleeff J, Guweidhi A, et al. RUNX3 expression in primary and metastatic pancreatic cancer. J Clin Pathol 2004;57:294–299PMID : 14990603.



12. Al-Sukhun S, Hussain M. Molecular biology of transitional cell carcinoma. Crit Rev Oncol Hematol 2003;47:181–193PMID : 12900011.


13. Orntoft TF, Wolf H. Molecular alterations in bladder cancer. Urol Res 1998;26:223–233PMID : 9759995.


14. Hwang KT, Han W, Bae JY, et al. Downregulation of the RUNX3 gene by promoter hypermethylation and hemizygous deletion in breast cancer. J Korean Med Sci 2007;22(suppl):S24–S31PMID : 17923751.



15. Xiao WH, Liu WW. Hemizygous deletion and hypermethylation of RUNX3 gene in hepatocellular carcinoma. World J Gastroenterol 2004;10:376–380PMID : 14760761.



16. Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 2002;21:5427–5440PMID : 12154405.


17. Guo WH, Weng LQ, Ito K, et al. Inhibition of growth of mouse gastric cancer cells by Runx3, a novel tumor suppressor. Oncogene 2002;21:8351–8355PMID : 12447699.


18. Cohen MM Jr. TGF beta/Smad signaling system and its pathologic correlates. Am J Med Genet A 2003;116A:1–10PMID : 12476444.

Figure 1
Combined bisulfite restriction analysis result of Runt-related transcription factor 3 in thyroid cancer cell lines. The upper band represents unmethylated polymerase chain reaction (PCR) product. The lower cut band represents methylated PCR products. NC, negative control.
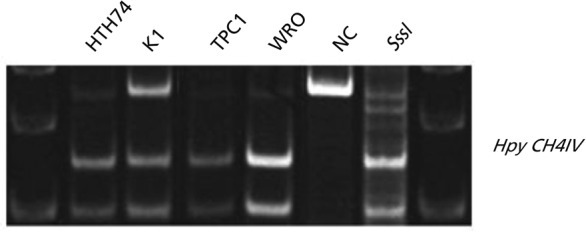
Figure 2
Combined bisulfite restriction analysis result of Runt-related transcription factor 3 in thyroid cancer cell tissues. The upper band represents unmethylated polymerase chain reaction (PCR) product. The lower cut band represents methylated PCR products. C, papillary thyroid cancer; H, Hurthle cell adenoma; SssI, positive control from SssI methylase-treated normal DNA; NC, negative control from DNA from the blood of normal healthy women.




 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print