|
 |
| Korean J Intern Med > Volume 38(6); 2023 > Article |
|
Abstract
Background/Aims
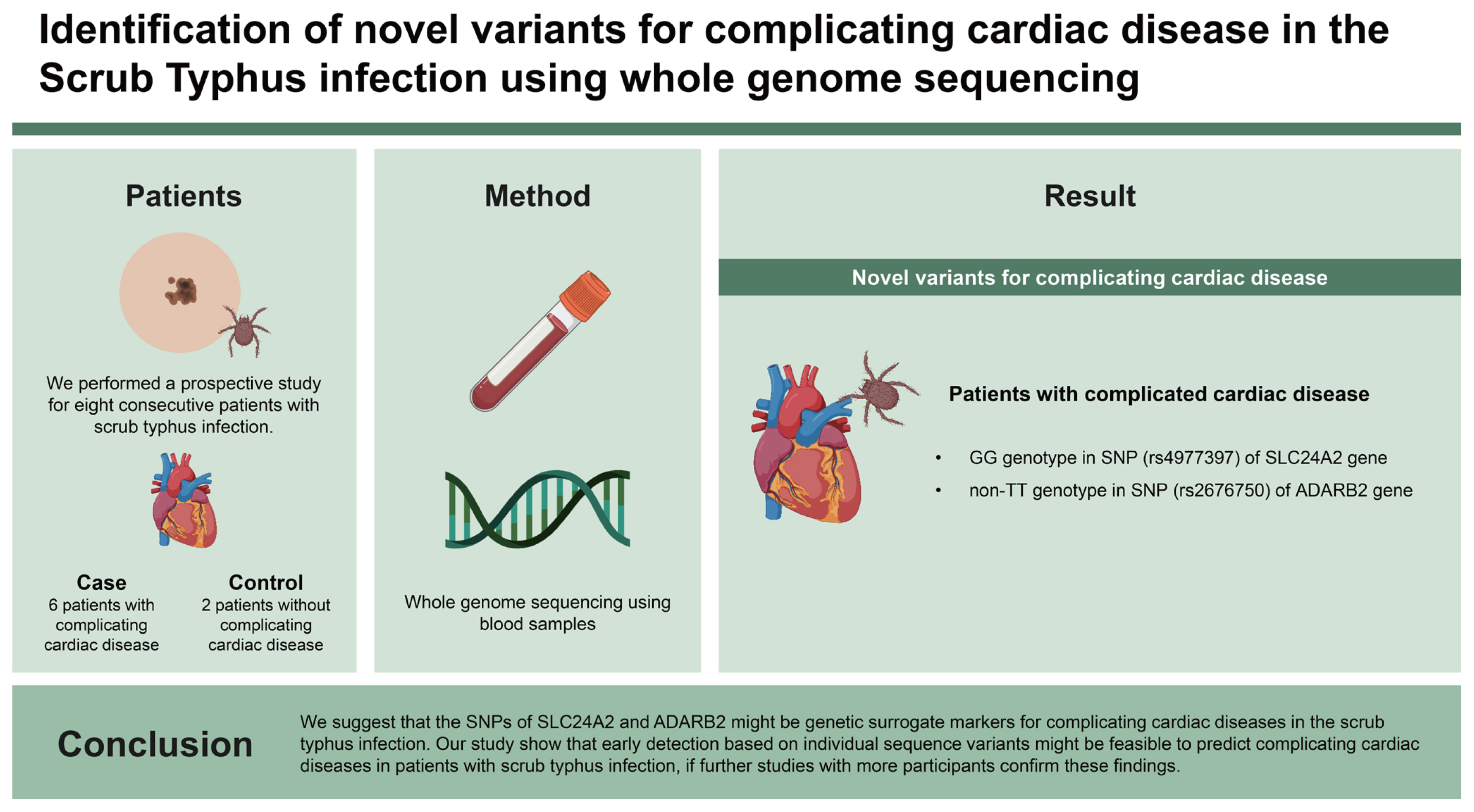
Scrub typhus infection has been known to complicate cardiovascular diseases mainly attributing to high mortality. Genetic susceptibility loci for complicating cardiac diseases such as atrial fibrillation, heart failure, and ischemic heart disease identified by genomic study have been limited in scrub typhus infection. Therefore, we investigated the genetic novel variants predicting complicating cardiac diseases in patients with confirmed scrub typhus infection using whole genome sequencing.
Methods
We performed a prospective study for eight consecutive patients with scrub typhus infection. During follow-up, six cases were clinically diagnosed with complicating cardiac diseases and two controls without complicating cardiac diseases. The whole genomes of the all patients were sequenced, and the individual sequence variants were compared between case and control patients. Variant genotypes were compared and identified as a single nucleotide polymorphism (SNP) of the different genotype distributions between six cases and two controls.
Results
The GG genotype in SNP (rs4977397) of solute carrier 24 family member 2 (SLC24A2) gene and non-TT genotype in SNP (rs2676750) of adenosine deaminase, RNA specific, B2 (ADARB2) gene were distinctively found in the case patients with complicated cardiac disease, compared with control patents in the scrub typhus infection.
Conclusions
We suggest that the SNPs of SLC24A2 and ADARB2 might be genetic surrogate markers for complicating cardiac diseases in the scrub typhus infection. Our study show that early detection based on individual sequence variants might be feasible to predict complicating cardiac diseases in patients with scrub typhus infection, if further studies with more participants confirm these findings.
Scrub typhus is a well-known endemic seasonal infectious disorder confined to Southeastern Asia and Western Pacific rim [1–3] and complicating cardiovascular adverse outcomes remain still under-recognized [4,5]. Even though the most of patients with scrub typhus infection can be cured with conventionally infectious control [6], as regard to complicated cardiovascular disease in the scrub typhus infection, the overall mortality rate has been reported from 16.7% to 30% which might be attributed to cardiovascular complication [7]. In addition, scrub typhus infection-induced cardiovascular complication has been recently reported as poor prognosis factor which have been recognized atrial fibrillation (AF) as clinical surrogate marker and associated with cardiovascular complication [8,9]. In addition, Genome-wide association studies (GWAS) have been utilized to assess the pathologically genetic connection of cardiovascular diseases and genetic susceptibility in the development of cardiovascular complication [2,3]. A recent GWAS was mostly performed and identified approximately several common genetic susceptibility loci for AF, heart failure (HF), and ischemic heart disease (IHD). Genetic susceptibility loci for complicating cardiac diseases such as AF, HF, and IHD identified by genomic study have been limited in scrub typhus infection. Therefore, we investigated the genetic novel variant/marker predicting complicating cardiac diseases such as AF, HF, and IHD in patients with confirmed scrub typhus infection using whole genome sequencing.
Consecutive eight patients with scrub typhus infection participated in this study. This study were approved by the Institutional Review Board in Chung-Ang University Hospital (2109-002-476) and Eulji University Hospital (EMC 2017-10-006) and adhered to the principles of the Declaration of Helsinki. The whole blood samples were corrected in ethylene-diamine-tetraacetic acid (EDTA) tube for genomic DNA extraction.
Genomic DNA was extracted from eight whole blood samples with ExgeneTM Blood SV kit (GeneAll, Seoul, Korea), following the manufacturer’s protocol (Blood SV mini exgene). Genomic DNA was analyzed by Qubit fluorometer dsDNA assay Kit (Invitrogen, Carlsbad, CA, USA) as well as Infinite F200 Pro NanoQuant (TECAN, Männedorf, Switzerland) to verify the quality (O.D. 260/280 ratio is 1.8–2.0 and O.D. 260/230 ratio greater than 1.6) and quantity (1 μg for library construction).
Genomic DNA for input into the TruSeq Nano DNA protocol (Truseq Nano DNA Library Prep Reference Guide) was quantified and diluted to 2 ng/μL. Twenty-five microlitres of genomic DNA were sheared using an S2 Ultrasonicator (Covaris, Woburn, MA, USA) using the settings as for the TruSeq DNA protocol (Truseq Nano DNA Library Prep Reference Guide). Library preparation was performed according to the manufacturer’s instructions. Adaptor enrichment was performed using eight cycles of enrich DNA fragments (polymerase chain reaction) according to the manufacturer’s instructions. The final products were quantified using the Agilent TapeStation 4200 HSD1000 screen tapes (Agilent Technologies, Santa Clara, CA, USA) and KAPA Library Quantification Kit (KK4824; Kapa Biosystems, Vienna, Austria). The individual samples were pooled and sequenced on the Illumina NovaSeq6000 (Illumina, San Diego, CA, USA) with 150 bp paired-end by following the manufacturer’s protocols. Image analysis were performed using the NovaSeq6000 control Software (version 1.3.1; Illumina) and the output data was demultiplexed with bcl2fastq (version 2.20; Illumina).
The quality of the reads was checked using fastQC (version 0.11.7; Illumina), helping the understanding for the basic quality for sequence quality score, GC contents, N contents, length distribution, and duplication level. After checking the read quality, the low-quality base below Q20 (accuracy 99%) were trimmed using Trimmomatic (version 0.36). High quality reads were then aligned to the human reference genome hg19 using BWA (version 0.7.17) [10] with minimum seed length of 45. After the alignment of the reads to reference genome, the duplicated reads were further removed using MarkDuplicates in GATK (version 4.0.11.0)
For the read alignment, whole chromosome regions were split for each chromosome using samtools (version 1.8) [11,12]. Then, Base Quality Score Recalibration (BQSR) process was done to adjust the quality score using BaseRecalibrator in GATK (version 4.0.11.0) [13]. For the realigned and recalibrated reads, variants were called using HaplotypeCaller GATK (version 4.0. 11.0) [12,14].
Regardless of stroke, we classified the control group as patients without cardiac disease and the case group as patients with cardiac disease. Scrub typhus induced cardiac complication was defined as new-onset AF, acute HF and IHD. New-onset AF was defined with a diagnosis code of ICD-10 I48 (paroxysmal AF) within 30 days of the index date without previous history of AF. New-onset acute HF was defined with a diagnosis code of ICD-10 I40 (acute myocarditis), I30 (acute pericarditis), or I50 (HF) within 30 days of the index date. New-onset IHD was defined 1) with a diagnosis code of ICD-10 I21 (acute myocardial infarction) or I20 (angina pectoris) within 30 days of the index date, and 2) treatment with coronary bypass graft surgery, primary coronary intervention, or thrombolytic agents.
A total of eight patients with scrub typhus infection (see Table 1) were subjected to Whole Genome Sequencing analysis. The amount of sequence generated from each sample was about 96 to 148 Gb, producing a sequence that is 30× or more of the total genomic DNA amount. Compared to the human reference genome sequence, a total of 4.28 million to 4.39 million variant sequences were detected. Among them, about 3.4 million single nucleotide polymorphisms (SNPs) were detected, and about 35,000 mutations were located in the protein coding regions. In this study, among these SNPs, SNPs that have been reported to be related to cardiovascular disease through previous studies (see Table 2) were extracted and analyzed to see if there was a difference between the case and the control group.
Table 1 shows the clinical characteristics of all enrolled patients with scrub typhus infection. Case patients developed new-onset AF, HF or acute coronary syndrome in longer stay of intensive care units, compared with control patients. Table 2 shows the previously reported SNPs related with cardiovascular diseases. Among these SNPs, the rs4977397 (chromosome 9, n.202350004A>G), solute carrier 24 family member 2 (SLC24A2) gene was distinctively found in the case patients with complicated cardiac disease, compared with control patents in the scrub typhus infection. Table 3 shows SNP in case patients with scrub typhus infection among previously reported SNPs not related with cardiovascular disease. The rs2676750 (chromosome 10, c.101-99601A>G), non-TT genotype, adenosine deaminase, RNA specific, B2 (ADARB2) gene was distinctively found in the case patients with complicated cardiac disease in the scrub typhus infection. Finally, the GG genotype in SNP (rs4977397) of SLC24A2 and non-TT genotype in SNP (rs2676750) of ADARB2 in patients with cardiac complications after scrub typhus infection is illustrated in Table 4.
This study presents genetic surrogate markers for cardiac complications associated with the scrub typhus infection and it is the first trial conducting a risk-analysis of the occurrence of cardiac adverse events after the infection. In the present study, GG genotype in rs4977397 of SLC24A2 and non-TT genotype in rs2676750 of ADARB2 were distinctively found in the patients with complicating cardiac disease compared with patients without complicating cardiac disease in the scrub typhus infection. This is therefore the first study depicting the variants of SLC24A2 and ADARB2 genes implicated in cardiac complications due to scrub typhus infection.
In the present study, newly developed clinical AF was observed in patients under case group, with some combination of HF and IHD compared with control patients (Table 1), which was also demonstrated in the Korean national data analysis in the scrub typhus infection [9]. It is known that new-onset AF is significantly associated with all-cause death in critically ill patients with other common infectious diseases and has the incidence risk of 2.2-fold for 1-year death compared to those without new-onset AF [15]. The Korean national data also shows that new-onset AF occurred in the 7.7% of all patients in the intensive care unit and has an incidence risk of 4.5-fold for 3-month mortality compared with those without new-onset AF and cardiovascular complication has been developed within only few days of index diagnosis of scrub typhus infection [9]. The reason for complicating cardiovascular diseases could be explained by possible pathophysiology of scrub typhus infection itself which initiates at the site of skin inoculation, evolves into regional lymphadenopathy and spreads to vasculitis with subsequent target organ damage [16]. Subsequently, cardiovascular inflammation associated with the scrub typhus infection can lead to cardiovascular complications. Unlike other infectious diseases, evidence to date has shown a robust association between scrub typhus infection and cardiovascular complications.
The Nav1.5, cardiac-type voltage-dependent Na+ channel (SCN5A) and KV11.1, delayed rectifier K+ channels (KCNH2) have previously been implicated in the pathophysiology including AF and cardiac conduction disorders through international GWAS [17,18]. Another members of the Na+/Ca2+/K+ exchangers gene family, cone/neuronal K+-Dependent Na+/Ca2+ Exchanger Isoform 2 (NCKX2), and rs10738554 of SLC24A2 gene, were previously associated with high blood pressure, which regulate to transport sodium, potassium, and calcium ions for homeostasis in the cardiovascular system [19,20]. However, SNP (rs4977397) of SLC24A2 has not been reported in patients with cardiac complications after scrub typhus infection. The rs4977397 SNP is located approximately 40 kb upstream of the SLC24A2 gene and has been reported to increase the risk of AF by approximately 1.04 times (p value = 9.0 × 10−9) when carrying the A allele [18].
ADARB2 is a member of the double-stranded RNA adenosine deaminase family of RNA-editing enzymes [21]. To date, only a few association studies have investigated SNPs in the RNA editing genes ADARB1 and ADARB2. ADARB2 gene-smoking interactions affect hypertension [22]. ADARB2, rs2805533 is located in intron 7 of ADARB2 and rs2805533 induces the splicing abnormality of ADARB2 and modulates the function of ADARB2 [23]. However, the SNP (rs2676750) of ADARB2 associated with cardiac diseases has net been reported, yet. The rs2676750 SNP is located in intron 1 of the ADARB2 gene, and there has been no previous paper reported on the SNP.
Both SNPs (rs4977397 and rs2676750) are not located in the protein-coding region, so their effect on protein function is thought to be very small. Further research on the specific functions of SLC24A2 and ADARB2 in the heart is needed.
The present study is the first to demonstrate that scrub typhus infection could be genetically linked to cardiac complications through the co-segregation between genetic markers (SNPs of SCL24A2 and ADARB2) and cardiac complications. This extends our understanding of the biological connection between the genetic loci and cardiac complications in scrub typhus infection. However, a limitation of this study is that the number of patients studied is too small to conclude on the clinical significance. Further studies with more participants are required to confirm and validate these findings.
We suggest that GG genotype in SNP (rs4977397) of SLC24A2 gene and non-TT genotype in SNP (rs2676750) of ADARB2 gene might be genetic surrogate markers for complicating cardiac disease in the scrub typhus infection. Our study shows that early detection based on individual sequence variants might be feasible for predicting complicating cardiac diseases in patients with scrub typhus infection, if further studies with more participants confirm these findings.
1. SNP (rs4977397) of SLC24A2 gene and SNP (rs2676750) of ADARB2 gene might be novel variants for complicating cardiac disease in patients with scrub typhus infection.
2. The genetic novel variants using whole genome sequencing might predict complicating cardiac diseases in patients with confirmed scrub typhus infection.
Notes
CRedit authorship contributions
Ki-Woon Kang: conceptualization, methodology, resources, investigation, data curation, formal analysis, validation, writing - original draft, funding acquisition; Kyung-Won Hong: data curation, validation, software, visualization; Seong-Kyu Lee: conceptualization, writing - review & editing, supervision
Table 1
Clinical baseline characteristics in all patients with scrub typhus infection
Table 2
Previously reported SNPs related with cardiovascular diseases
| Chr | SNP ID | Gene symbol | HGVS | Ref | Alt | Control patients | Case patients | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||
| 1 | 2 | 1 | 2 | 3 | 4 | 5 | 6 | ||||||
| 1 | rs660240 | CELSR2 | c.*1167T>C | T | C | CC | CC | TC | CC | CC | CC | CC | CC |
|
|
|||||||||||||
| 1 | rs608930 | GORAB | n.170617306G>T | G | T | GT | GT | GT | GT | TT | GT | GT | GT |
|
|
|||||||||||||
| 1 | rs503706 | PRRX1 | c.241+1484T>C | T | C | TC | TC | TC | CC | CC | TC | TC | TC |
|
|
|||||||||||||
| 2 | rs515135 | APOB | n.21286057T>C | T | C | CC | CC | CC | CC | TC | CC | CC | CC |
|
|
|||||||||||||
| 2 | rs6544713 | ABCG8 | c.322+431T>C | T | C | CC | CC | CC | CC | CC | CC | CC | CC |
|
|
|||||||||||||
| 2 | rs2252641 | TEX41 | n.464−26926T>C | T | C | CC | TC | CC | TC | CC | CC | CC | CC |
|
|
|||||||||||||
| 2 | rs840616 | CALCRL | n.188196469T>C | T | C | CC | CC | CC | CC | CC | CC | CC | CC |
|
|
|||||||||||||
| 2 | rs3820888 | SPATS2L | c.18+6210T>C | T | C | TC | TC | TC | TC | CC | CC | TC | TC |
|
|
|||||||||||||
| 3 | rs6790396 | SCN10A | c.2281−1533G>C | C | G | GG | GG | GG | GG | GG | GG | CG | GG |
|
|
|||||||||||||
| 3 | rs17005647 | FRMD4B | c.162+28798G>A | C | T | TT | CT | CT | CT | CT | TT | TT | TT |
|
|
|||||||||||||
| 3 | rs667920 | STAG1 | c.3066−1267C>A | G | T | TT | TT | TT | TT | TT | TT | GT | TT |
|
|
|||||||||||||
| 3 | rs4266144 | CCNL1 | n.156852592C>G | C | G | GG | CG | CG | CG | GG | CG | GG | CG |
|
|
|||||||||||||
| 3 | rs60902112 | XXYLT1 | c.786−10013G>A | C | T | CT | CT | TT | CT | CT | CT | TT | CT |
|
|
|||||||||||||
| 4 | rs3960788 | SLC9B1 | c.−1−2749A>G | T | C | CC | TC | TC | TC | TC | TC | TC | TC |
|
|
|||||||||||||
| 4 | rs6847935 | PITX2 | n.111696651A>T | A | T | AT | AT | TT | TT | AT | AT | AT | TT |
|
|
|||||||||||||
| 5 | rs6596717 | EFNA5 | n.106427609C>A | C | A | CA | CA | CA | AA | CA | CA | CA | AA |
|
|
|||||||||||||
| 5 | rs2012809 | SLC27A6 | n.128190363A>G | A | G | GG | GG | GG | GG | GG | GG | GG | GG |
|
|
|||||||||||||
| 6 | rs1307274 | NUDT3 | n.34240576T>G | T | G | TG | GG | GG | GG | GG | GG | GG | TG |
|
|
|||||||||||||
| 6 | rs6905288 | VEGFA | c.*6574G>A | G | A | GA | GA | AA | GA | AA | GA | AA | AA |
|
|
|||||||||||||
| 6 | rs1591805 | CENPW | n.126717064A>G | A | G | GG | GG | GG | GG | GG | GG | GG | GG |
|
|
|||||||||||||
| 7 | rs11509880 | TMEM106B | c.282−1941G>A | G | A | AA | GA | GA | AA | AA | GA | GA | AA |
|
|
|||||||||||||
| 7 | rs6462078 | CREB5 | n.28413187C>A | C | A | AA | AA | AA | AA | AA | AA | AA | AA |
|
|
|||||||||||||
| 9 | rs4977397 | SLC24A2a) | n.20235004A>G | A | G | AG | AG | GG | GG | GG | GG | GG | GG |
|
|
|||||||||||||
| 10 | rs61848342 | CAMK1D | n.12303813T>C | T | C | CC | TC | CC | TC | TC | TC | TC | TC |
|
|
|||||||||||||
| 10 | rs7096385 | SIRT1 | c.943−1666T>C | T | C | CC | TC | CC | TC | TC | TC | CC | CC |
|
|
|||||||||||||
| 10 | rs10749053 | RBM20 | c.2551−3135T>C | T | C | CC | CC | CC | TC | CC | CC | CC | CC |
|
|
|||||||||||||
| 11 | rs949078 | SORL1 | n.121629007C>T | C | T | TT | CT | TT | TT | TT | CT | TT | CT |
|
|
|||||||||||||
| 12 | rs4766578 | ATXN2 | c.3317−1852A>T | T | A | AA | AA | AA | AA | AA | AA | AA | AA |
|
|
|||||||||||||
| 12 | rs6560886 | FBRSL1 | c.1585−695T>C | T | C | CC | CC | CC | CC | CC | CC | CC | TC |
|
|
|||||||||||||
| 14 | rs2738413 | SYNE2 | c.19056+237A>G | A | G | AG | GG | GG | AG | AG | AG | GG | GG |
|
|
|||||||||||||
| 14 | rs1152591 | SYNE2 | c.19057−64A>G | A | G | AG | GG | GG | AG | AG | AG | GG | GG |
|
|
|||||||||||||
| 14 | rs10873299 | IRF2BPL | n.77426711A>G | A | G | GG | AG | AG | AG | AG | GG | GG | AG |
|
|
|||||||||||||
| 16 | rs2286466 | RPS2 | c.261T>C | A | G | GG | GG | GG | GG | GG | GG | AG | GG |
|
|
|||||||||||||
| 19 | rs12976411 | ZNF507 | c.*8031A>T | A | T | AT | AT | AT | TT | AT | TT | AT | TT |
Table 3
SNP in case patients with scrub typhus infection among previously reported SNPs not related with cardiovascular disease
| Chr | SNP ID | Gene symbol | HGVS | Ref | Alt | Control patients | Case patients | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||
| 1 | 2 | 1 | 2 | 3 | 4 | 5 | 6 | ||||||
| 10 | rs2676750 | ADARB2 | c.101−99601A>G | T | C | TT | TT | CC | CC | CC | CC | TC | CC |
REFERENCES
1. Sittiwangkul R, Pongprot Y, Silviliarat S, Oberdorfer P, Jittamala P, Sirisanthana V. Acute fulminant myocarditis in scrub typhus. Ann Trop Paediatr 2008;28:149–154.


2. Clauss S, Sinner MF, Kääb S. Genome-wide association studies revealing the heritability of common atrial fibrillation: is bigger always better? Circ Cardiovasc Genet 2017;10:e002005.

3. Weng LC, Choi SH, Klarin D, et al. Heritability of atrial fibrillation. Circ Cardiovasc Genet 2017;10:e001838.


4. Walker DH. Scrub typhus - scientific neglect, ever-widening impact. N Engl J Med 2016;375:913–915.


5. Weitzel T, Dittrich S, López J, et al. Endemic scrub typhus in South America. N Engl J Med 2016;375:954–961.


6. Peter JV, Sudarsan TI, Prakash JA, Varghese GM. Severe scrub typhus infection: clinical features, diagnostic challenges and management. World J Crit Care Med 2015;4:244–250.



7. Lee CS, Hwang JH, Lee HB, Kwon KS. Risk factors leading to fatal outcome in scrub typhus patients. Am J Trop Med Hyg 2009;81:484–488.


8. Chin JY, Kang KW, Moon KM, Kim J, Choi YJ. Predictors of acute myocarditis in complicated scrub typhus: an endemic province in the Republic of Korea. Korean J Intern Med 2018;33:323–330.




9. Jang SY, Kang KW, Kim JH, et al. New-onset atrial fibrillation predicting for complicating cardiac adverse outcome in scrub typhus infection. Clin Cardiol 2019;42:1210–1221.




10. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760.




11. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303.



12. Danecek P, Bonfield JK, Liddle J, et al. Twelve years of SAMtools and BCFtools. Gigascience 2021;10:giab008.




13. Carey VJ, Ramos M, Stubbs BJ, et al. Global alliance for genomics and health meets bioconductor: toward reproducible and agile cancer genomics at cloud scale. JCO Clin Cancer Inform 2020;4:472–479.



14. Li H, Handsaker B, Wysoker A, et al.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–2079.




15. Arrigo M, Ishihara S, Feliot E, et al. New-onset atrial fibrillation in critically ill patients and its association with mortality: a report from the FROG-ICU study. Int J Cardiol 2018;266:95–99.


16. Allen AC, Spitz S. A comparative study of the pathology of scrub typhus (tsutsugamushi disease) and other rickettsial diseases. Am J Pathol 1945;21:603–681.


17. Olson TM, Michels VV, Ballew JD, et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA 2005;293:447–454.



18. Roselli C, Chaffin MD, Weng LC, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet 2018;50:1225–1233.


19. Li XF, Kraev AS, Lytton J. Molecular cloning of a fourth member of the potassium-dependent sodium-calcium exchanger gene family, NCKX4. J Biol Chem 2002;277:48410–48417.


20. Kodaman N, Sobota RS, Asselbergs FW, et al. Genetic effects on the correlation structure of CVD risk factors: exome-wide data from a ghanaian population. Glob Heart 2017;12:133–140.



21. Hogg M, Paro S, Keegan LP, O’Connell MA. RNA editing by mammalian ADARs. Adv Genet 2011;73:87–120.





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print