LPL gene Pvu II polymorphism and hypertriglyceridemia: a meta-analysis involving 1,640 subjects
Article information
Abstract
Background/Aims
Although lipoprotein lipase (LPL) gene Pvu II polymorphism has been associated with an increased risk of hypertriglyceridemia (HT), there is no clear consensus within the scientific community.
Methods
A meta-analysis of 1,640 subjects from six individual studies was conducted to better elucidate the potential relationship between the LPL gene Pvu II polymorphism and HT within the Chinese population. Pooled odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) were evaluated by using fixed effect models.
Results
Our analysis indicated a significant association between LPL gene Pvu II polymorphism and HT within the Chinese population under allelic (OR, 1.550; 95% CI, 1.320 to 1.830; p = 1.158 × 10-7), recessive (OR, 0.540; 95% CI, 0.390 to 0.750; p = 0.0002), dominant (OR, 1.889; 95% CI, 1.501 to 2.377; p = 5.960 × 10-8), homozygous (OR, 2.167; 95% CI, 1.531 to 3.067; p = 1.242 × 10-5), heterozygous (OR, 1.810; 95% CI, 1.419 to 2.309; p = 1.842 × 10-6), and additive genetic models (OR, 1.553; 95% CI, 1.320 to 1.828; p = 1.158 × 10-7).
Conclusions
Because LPL gene Pvu II restriction fragment length polymorphism polymorphism was associated with an elevated risk of HT, the P+ allele carriers of the LPL gene might be predisposed to HT.
INTRODUCTION
Hyperlipidemia (HL) is an umbrella term referring to several acquired or genetic disorders that result in high level of lipids circulating in the blood. The increased prevalence of Western diets and sedentary lifestyles contributed to increasing morbidities and mortalities associated with HL, but the complex interactions between hereditary and environments makes the disease difficult to treat.
One of the disorders categorized under HL is hypertriglyceridemia (HT), which, more specifically, describes elevated levels of triglycerides (TGs), the most abundant lipid in the human body [1]. HT is strongly associated with atherosclerosis, even in the absence of hypercholesterolemia, and diabetes mellitus. Contemporary research also identifies HT as one of the most potent risk factors for coronary artery disease [2]. These findings sparked the interest of many researchers into better understanding the hereditary and environmental factors that contribute to development of HT among patients.
Lipoprotein lipase (LPL) is an area of particular interest. LPL plays a key role in TG metabolism by catalyzing digestion of TGs in chylomicron emulsions and very low density lipoprotein (VLDL)’s into free fatty acids and glycerol to meet the energy expenditure and storage needs for a variety of high-energy tissues (adipocytes, cardiomyocytes, skeletal muscle cells, macrophages, etc) [3]. The importance of LPL in HT is further accentuated when we look at patients. Patients with HT always exhibit abnormal apolipoprotein levels with the significantly elevated plasma TG and VLDL level.
It has been postulated that variation within the genetic material coding this protein may also contribute to HT. The LPL gene, located in 8p22, is comprised of 10 exons and nine introns. The Pvu II polymorphism (rs285) is caused by the C-T base transition in the 6th intron. Although this polymorphism does not affect the structure of the encoded protein, it could still affect general TG esterlysis by altering the gene expression and regulation through linkage disequilibrium [4].
Many studies seeking to elucidate the relationship between this gene polymorphism and HT have been conducted, but they fail to present a clear consensus on the subject. This may be due to the small samples sizes of the each study individually. For example, while Fan et al. [5] and Wang et al. [6] both found that the P+P+ genotype in the LPL gene was associated with HT in separate Chinese populations, but Huang et al. [7] found no genetic risk among HT subjects. Nevertheless, only 328, 464, and 310 sample size were included in the three individual studies respectively (Table 1). Our current meta-analysis seeks to combine the information gathered thus far and present a clear representation of the combined results.
METHODS
Publication search and inclusion criteria
Our primary search was executed through a search of the Wan Fang database, PubMed, the China National Knowledge Infrastructure, the VIP database, the China Biological Medicine Database, Embase, and the Web of Science using the terms “LPL,” “Pvu II,” “hypertriglyceridemia,” and “polymorphism.” Our initial search yielded papers published between 1999 and 2007 with the most recent update on May 28, 2016.
Studies selected for our meta-analysis had to meet inclusion criteria as follows. Studies selected must (1) evaluate the association of HT and LPL gene Pvu II polymorphism, (2) diagnose HT as defined by TG ≥ 2.5 mmol/L, and (3) have control groups that follow Hardy-Weinberg equilibrium (HWE). Results of the studies must also be published in official journals as either cohort or case-control studies.
Data extraction
The data was drawn out using a standardized protocol by three investigators. Two were responsible for identifying duplicate studies with the third serving as an arbitrator to settle any differences. Those studies that deviated from the inclusion criteria, repeatedly published, or did not provide sufficient data were removed. Similar data sets presented in different publications by the same author group were used once in our study. The listed data table should include such items as the first author’s name, publication year, region, genotyping method, matching criteria, the genotype number in the HT and control groups, and sample size of cases and controls.
Statistical analyses
Six genetic models were used in the present meta-analysis: allelic (P+ allele distribution frequency), recessive (P–P– vs. P+P+ and P+P–), dominant (P+P+ vs. P–P– and P+P–), homozygous (P+P+ vs. P–P–), heterozygous (P+P+ vs. P+P–), and additive (P+ vs. P–). The odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) were used to compare the relationship of LPL gene Pvu II polymorphism and HT. The heterogeneity among the studies was calculated by using the chi-square-based Q-tests with the significance set at p < 0.05 level [8]. The fixed-effect model (the Mantel-Haenszel method) was used [9] because no heterogeneity among the included studies. The pooled OR was assessed by Z test with the significance set at p < 0.05 level.
HWE was evaluated by using the Fisher exact test with the significance set at p < 0.05 level. Potential publication bias was assessed using a funnel plot. Funnel plot symmetry was evaluated by using the Egger’s linear regression test on a natural log scale of the OR with the significance set at p < 0.05 level [10]. Statistical analyses were performed by using Stata version 12.0 (StataCorp., College Station, TX, USA) and Review Manager 5.0.
RESULTS
Studies and populations
All of information was abstracted from 515 HT cases and 1,125 controls (Table 1) [5-7,11-13]. Twelve papers were obtained through the retrieval process, among which six papers were included in the present meta-analysis. Among the six excluded studies, two papers were reviews and two papers deviated from the HWE [14,15], and two papers have nothing with LPL gene Pvu II polymorphism or HT.
Pooled analyses
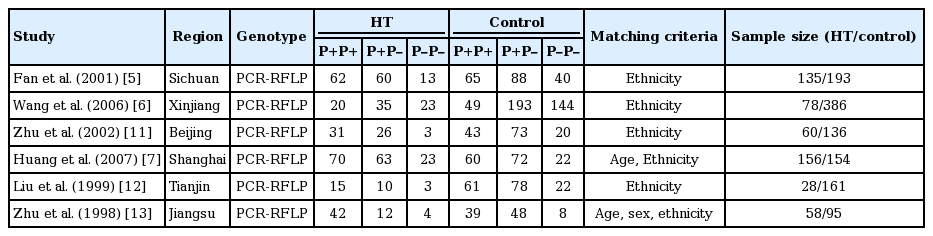
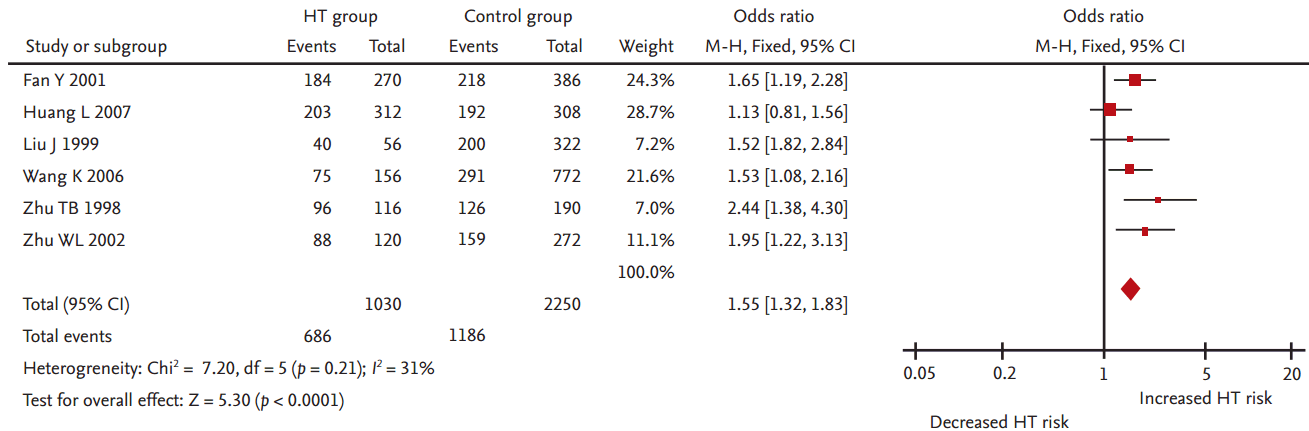
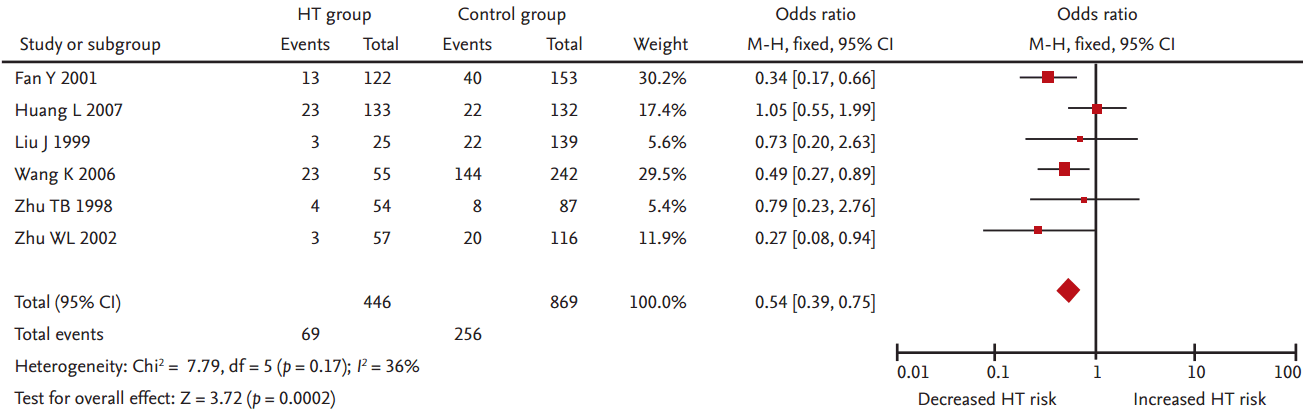
A significant association between LPL gene Pvu II polymorphism and HT was detected in the Chinese population under all genetic models: allelic (OR, 1.550; 95% CI, 1.320 to 1.830; p = 1.158 × 10–7), recessive (OR, 0.540; 95% CI, 0.390 to 0.750; p = 0.0002), dominant (OR, 1.889; 95% CI, 1.501 to 2.377; p = 5.960 × 10–8), homozygous (OR, 2.167; 95% CI, 1.531 to 3.067; p = 1.242 × 10–5), heterozygous (OR, 1.810; 95% CI, 1.419 to 2.309; p = 1.842 × 10–6), and additive genetic models (OR, 1.553; 95% CI, 1.320 to 1.828; p = 1.158 × 10–7) (Table 2, Figs. 1-6).

Summary of meta-analysis of association between LPL gene Pvu II polymorphism and HT in the Chinese population
Forest plot of hypertriglyceridemia (HT) associated with lipoprotein lipase (LPL) gene Pvu II polymorphism under an allelic genetic model (distribution of P+ allelic frequency of LPL gene Pvu II polymorphism). CI, confidence interval.
Forest plot of hypertriglyceridemia (HT) associated with lipoprotein lipase (LPL) gene Pvu II polymorphism under a recessive genetic model (P–P– vs. P+P+ + P+P–). CI, confidence interval.

Forest plot of hypertriglyceridemia associated with lipoprotein lipase (LPL) gene Pvu II polymorphism under a dominant genetic model (P+P+ vs. P–P– + P+P–). OR, odds ratio; CI, confidence interval.
Forest plot of hypertriglyceridemia associated with lipoprotein lipase (LPL) gene Pvu II polymorphism under a homozygous genetic model (P+P+ vs. P–P–). OR, odds ratio; CI, confidence interval.
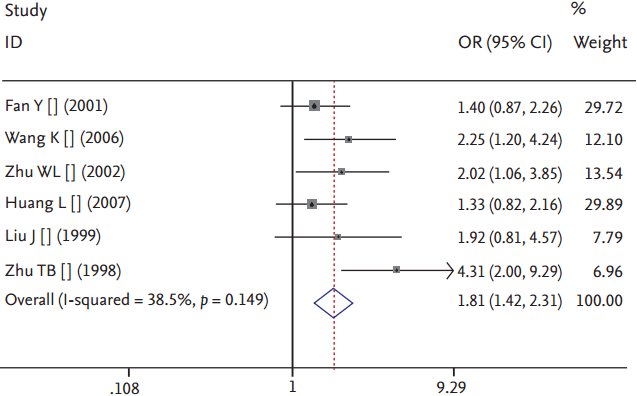
Forest plot of hypertriglyceridemia associated with lipoprotein lipase (LPL) gene Pvu II polymorphism under a heterozygous genetic model (P+P+ vs. P+P–). OR, odds ratio; CI, confidence interval.
Bias diagnostics
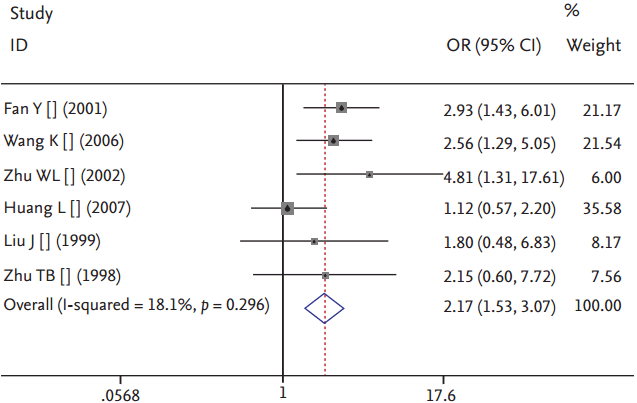
There was no visible publication bias in the funnel plot under the allelic genetic model (Fig. 7). Additionally, there was no significant difference in the Egger’s test yet, which suggested that no publication bias existed in the present meta-analysis by using additive genetic model (T = 1.57, p = 0.192).

The funnel plot for studies of the association of hypertriglyceridemia associated with lipoprotein lipase (LPL) gene Pvu II polymorphism under an allelic genetic model (distribution of P+ allelic frequency of LPL gene Pvu II polymorphism). The horizontal and vertical axis correspond to the odds ratio (OR) and confidence limits. SE, standard error.
DISCUSSION
Our meta-analysis found a significant relationship between the LPL gene Pvu II polymorphism and HT under all genetic models, which suggested that the gene polymorphism might predispose P+ allele carriers to HT.
However, the precise molecular mechanism behind this relationship remains unclear. Because the Pvu II restriction enzyme digestion site is located in an intron, its alteration would not directly affect the protein structure and its baseline activity. However, it could influence the LPL activity or regulate the gene expression by the chain effects [16]. Hayden et al. [17] also suggested that the base variation in the intron might interact with environmental factors such as excessive drinking, pregnancy, and diabetes mellitus to reduce the LPL gene expression and lead to the elevated TG level.
In 2002, Ukkola et al. [18] gave excessive dietary intervention to 12 pairs of healthy identical twins for 100 days and they found that the LPL gene Pvu II polymorphism was positively correlated with body mass index and LPL activity. Nicklas et al. [19] found that the LPL gene Pvu II polymorphism was not only associated with the LPL activity, but also with the serum lipid level.
Our meta-analysis improves on the presenting the current state of research on the topic. An admirable meta-analysis performed by Cui et al. [20] in 2010 also found the LPL gene Pvu II polymorphism to be significantly associated with HL. However, two studies with controls deviating from the HWE were included in their meta-analysis [20]. Hence, its results might not reach the objective standard of our presented work.
Even so, our current meta-analysis does not present a large-scale study necessary to fully understand the relationship between the LPL gene Pvu II polymorphism and HT. Our study also fails to take into account environmental factors that play a significant role in the development HT. It also fails to take into account the micro-effects of many other genes involved in lipid metabolism, transport, and storage (i.e., CRTC2 gene rs8450 polymorphism, MTTP 297 H polymorphisms) [21,22].
In conclusion, LPL gene Pvu II polymorphism was significantly associated with increased HT risk in the Chinese population. The persons with the P+ allele of LPL gene Pvu II polymorphism might be easy to suffer from HT. The significant association between them suggested that more studies clarifying more about this complex relationship would be valuable to developing our understanding of this subject.
KEY MESSAGE
1. Lipoprotein lipase (LPL) gene Pvu II restriction fragment length polymorphism was associated with an elevated risk of hypertriglyceridemia (HT).
2. The P+ allele carriers of the LPL gene might be predisposed to HT.
Notes
No potential conflict of interest relevant to this article was reported.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (NSFC 81100073 to Dr. Yan-yan Li), Excellent Young and Middle-Aged Teachers Assistance Program of Nanjing Medical University for Dr. Yan-yan Li (2013–2015, JX2161015034), Jiangsu Overseas Research & Training Program for University Prominent Young & Middle-aged Teachers and Presidents, and the Priority AHLemic Program Development of Jiangsu Higher Education Institutions (PAPD). Thank all our colleagues working in the Department of geriatrics, the First Affiliated Hospital of Nanjing Medical University.